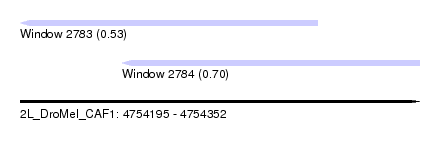
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,754,195 – 4,754,352 |
| Length | 157 |
| Max. P | 0.698337 |

| Location | 4,754,195 – 4,754,312 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.36 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -13.29 |
| Energy contribution | -13.29 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4754195 117 - 22407834 UAGUCAAACAUAAUUUGACCUAAGCCAAGGCAUAAGCAAAUGUGACAGGCUCCAGUCAGCCAUAAAAGCCU---UCUCAGCUAGAACACUCAUGGACACUUCCAUAGUCAUAGUCAUAGU ..((((((.....))))))....((....)).........((((((.((((......)))).....(((..---.....))).....(((.(((((....))))))))....)))))).. ( -24.90) >DroPse_CAF1 221284 100 - 1 UAGUCAAACAUAAUUUGACCUAAGCCAAGGCAUAAGCAAAUGUGACAGGCUCCAGUCAGGCAUAAAAGCCUGGGUCAC---------AC------ACAU-CCCACCGACUCCGACU---- .((((.......(((((.(.((.((....)).)).))))))(((((.(....)..((((((......)))))))))))---------..------....-......))))......---- ( -24.70) >DroSec_CAF1 182646 115 - 1 UAGUCAAACAUAAUUUGACCUAAGCCAAGGCAUAAGCAAAUGUGACAGGCUCCAGUCAGCCAUAAAAGCCU---UCUCAGCUAGAACACUCAAGGACAUUUCCACAGUCAUAGUCAUA-- ..((((((.....))))))....((....)).........((((((.((((......)))).....(((..---.....)))...........(((....)))...))))))......-- ( -21.90) >DroYak_CAF1 189230 115 - 1 UAGUCAAACAUAAUUUGACCUAAGCCAAGGCAUAAGCAAAUGUGACAGGCUCCAGUCAGCCAUAAAAGCCU---UCUCAGCUGGAACACUCAUGGCUAUUUCCAUAGUCAUAGACAUA-- ..((((((.....))))))(((.((.(((((....((....))....((((......))))......))))---)....)))))....((.((((((((....)))))))))).....-- ( -25.20) >DroAna_CAF1 201591 90 - 1 UAGUCAAACAUAAUUUGACCUAAGCCAAGGCAUAAGCAAAUGUGACAGGCUCCAGUCGGCCAUAAAAGCCU---C------------AC------ACAUUCCCA-GGUCCCG-------- ................(((((..((....))....(..((((((..(((((((....)).......)))))---.------------.)------)))))..))-))))...-------- ( -20.51) >DroPer_CAF1 229009 100 - 1 UAGUCAAACAUAAUUUGACCUAAGCCAAGGCAUAAGCAAAUGUGACAGGCUCCAGUCAGGCAUAAAAGCCUGGGUCUC---------AC------ACAU-CCCACCGACUCCGACU---- .((((...((((.((((.(.((.((....)).)).)))))))))...((....((((((((......))))(((....---------..------....-)))...))))))))))---- ( -24.40) >consensus UAGUCAAACAUAAUUUGACCUAAGCCAAGGCAUAAGCAAAUGUGACAGGCUCCAGUCAGCCAUAAAAGCCU___UCUC_________AC______ACAUUCCCACAGUCACAGACA____ ..((((......(((((.(.((.((....)).)).)))))).))))(((((...............)))))................................................. (-13.29 = -13.29 + 0.00)

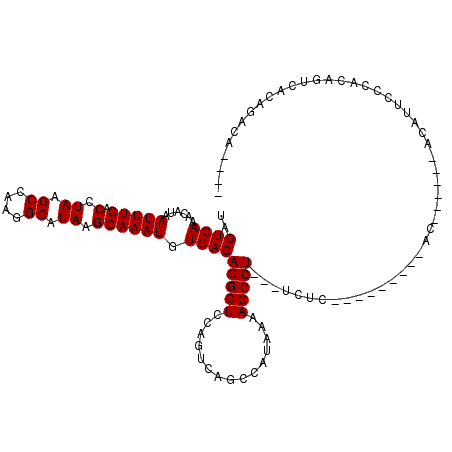
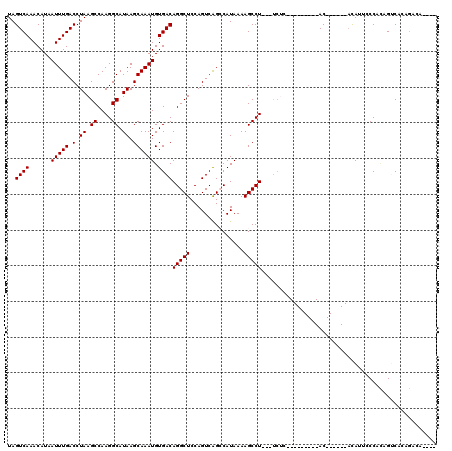
| Location | 4,754,235 – 4,754,352 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.48 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -28.31 |
| Energy contribution | -28.50 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4754235 117 - 22407834 UGUGCCCGGGGCUUUGUCCGAAAAUUUCGCAUGCCGUUUAUAGUCAAACAUAAUUUGACCUAAGCCAAGGCAUAAGCAAAUGUGACAGGCUCCAGUCAGCCAUAAAAGCCU---UCUCAG .......((((((((...........((((((...((((((.((((((.....))))))....((....))))))))..))))))..((((......))))...)))))))---)..... ( -31.10) >DroPse_CAF1 221306 118 - 1 UGUGCCCUGGGCUUUGUCCGAAAAUUUCGCAUGCCGUUUAUAGUCAAACAUAAUUUGACCUAAGCCAAGGCAUAAGCAAAUGUGACAGGCUCCAGUCAGGCAUAAAAGCCUGGGUCAC-- .(((.((..((((((...(((.....))).(((((((((((.((((((.....))))))....((....)))))))).....((((.(....).))))))))).))))))..)).)))-- ( -40.10) >DroEre_CAF1 191260 117 - 1 UGUGCUCGGGGCUUUGUCCGAAAAUUUCGCAUGCCGUUUAUAGUCAAACAUAAUUUGACCUAAGCCAAGGCAUAAGCAAAUGUGACAGGCUCCAGUCAGCCAUAAAAGCCU---UCUCAG .......((((((((...........((((((...((((((.((((((.....))))))....((....))))))))..))))))..((((......))))...)))))))---)..... ( -31.10) >DroYak_CAF1 189268 117 - 1 UGUGCCCGGGGCUUUGUCCGAAAAUUUCGCAUGCCGUUUAUAGUCAAACAUAAUUUGACCUAAGCCAAGGCAUAAGCAAAUGUGACAGGCUCCAGUCAGCCAUAAAAGCCU---UCUCAG .......((((((((...........((((((...((((((.((((((.....))))))....((....))))))))..))))))..((((......))))...)))))))---)..... ( -31.10) >DroAna_CAF1 201609 111 - 1 UGUGCCCUGGGCUUUGUCCGAA-AUUUCGCAUGCCGUUUAUAGUCAAACAUAAUUUGACCUAAGCCAAGGCAUAAGCAAAUGUGACAGGCUCCAGUCGGCCAUAAAAGCCU---C----- ((.(((((((((((.(((....-((((.((((((((((((..((((((.....)))))).)))))...)))))..))))))..))))))).))))..))))).........---.----- ( -32.00) >DroPer_CAF1 229031 118 - 1 UGUGCCCUGGGCUUUGUCCGAAAAUUUCGCAUGCCGUUUAUAGUCAAACAUAAUUUGACCUAAGCCAAGGCAUAAGCAAAUGUGACAGGCUCCAGUCAGGCAUAAAAGCCUGGGUCUC-- ...((((.(((((((...(((.....))).(((((((((((.((((((.....))))))....((....)))))))).....((((.(....).))))))))).)))))))))))...-- ( -37.30) >consensus UGUGCCCGGGGCUUUGUCCGAAAAUUUCGCAUGCCGUUUAUAGUCAAACAUAAUUUGACCUAAGCCAAGGCAUAAGCAAAUGUGACAGGCUCCAGUCAGCCAUAAAAGCCU___UCUC__ ........(((((((...........((((((...((((((.((((((.....))))))....((....))))))))..))))))..((((......))))...)))))))......... (-28.31 = -28.50 + 0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:46 2006