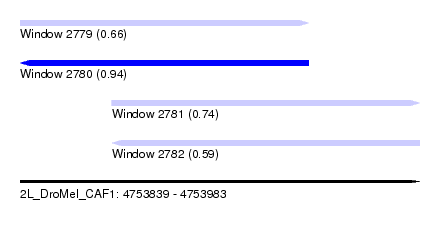
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,753,839 – 4,753,983 |
| Length | 144 |
| Max. P | 0.939116 |

| Location | 4,753,839 – 4,753,943 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.62 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -13.50 |
| Energy contribution | -14.06 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4753839 104 + 22407834 UAAAAUAUUAAAUGACCACAAUGUUUAAAGUGC--------AAUAA-------AUUUUCAUCGUGUGGCAUUAAUACCAUUGCCACACCACUGUUCCUGCCACUUUCUGCUGCUUGGCU ...............(((....((..(((((((--------(...(-------((.......((((((((..........))))))))....)))..)).))))))..))....))).. ( -21.30) >DroSec_CAF1 182306 104 + 1 UGAAAUAUUAAAUGGUCACAAUGUUUAAAGUGC--------AUUAA-------AUUCCCGGCGUGUGGCAUUUAUAUCGUUGCCACACCACUGUUCCUGCCACUUUUUGCUGCUUGGCU .............(((((....((..(((((((--------(...(-------((.....(.((((((((..........)))))))))...)))..)).))))))..))....))))) ( -24.80) >DroSim_CAF1 178236 104 + 1 UGAAAUAUUAAAUGGCCACAAUGUUUAAAGUGC--------AAUAA-------AUUUCCGGCGUGUGGCAUUAAAAUCGUUGCCACACCACUGUUCCUGCCACUUUUUGCUGCUUGGCU .............(((((....((..(((((((--------(...(-------((.....(.((((((((..........)))))))))...)))..)).))))))..))....))))) ( -27.50) >DroEre_CAF1 190889 103 + 1 UU-AAUAUUAAAUAGCCGCAAUGUUUAAACUGC--------AUUAA-------AUUUCAAGUGUAUAACAUUAAUAUCGUUGCCGCACCACUGUUCCUGCCACCUUUUGCUGCUCGGAU ..-...........((.((((((.((((..(((--------(((..-------......)))))).....))))...)))))).))....(((.....((........))....))).. ( -14.80) >DroYak_CAF1 188877 117 + 1 -G-AACAUUUUAGUACAGCAGUUUUCAGAUUGAAAGUUUUCGUUAAGUCACAGAUUUUAAGUGUGUGGCAUUAACAUCGUUGCCACACCACUGUUCGUGGCACUUUUUGCUGCUGUGAU -.-..........(((((((((((((.....))))...........(((((.((.....(((((((((((..........))))))).))))..))))))).......))))))))).. ( -36.60) >consensus UGAAAUAUUAAAUGGCCACAAUGUUUAAAGUGC________AUUAA_______AUUUCCAGCGUGUGGCAUUAAUAUCGUUGCCACACCACUGUUCCUGCCACUUUUUGCUGCUUGGCU ...............(((....((..((((((..............................((((((((..........))))))))((.......)).))))))..))....))).. (-13.50 = -14.06 + 0.56)


| Location | 4,753,839 – 4,753,943 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.62 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -15.91 |
| Energy contribution | -16.75 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4753839 104 - 22407834 AGCCAAGCAGCAGAAAGUGGCAGGAACAGUGGUGUGGCAAUGGUAUUAAUGCCACACGAUGAAAAU-------UUAUU--------GCACUUUAAACAUUGUGGUCAUUUAAUAUUUUA .((......))...(((((((....((((((((((((((..........))))))))(((((....-------)))))--------..........)))))).)))))))......... ( -27.00) >DroSec_CAF1 182306 104 - 1 AGCCAAGCAGCAAAAAGUGGCAGGAACAGUGGUGUGGCAACGAUAUAAAUGCCACACGCCGGGAAU-------UUAAU--------GCACUUUAAACAUUGUGACCAUUUAAUAUUUCA .((((............)))).((.((((((((((((((..........))))))))........(-------((((.--------.....)))))))))))..))............. ( -26.20) >DroSim_CAF1 178236 104 - 1 AGCCAAGCAGCAAAAAGUGGCAGGAACAGUGGUGUGGCAACGAUUUUAAUGCCACACGCCGGAAAU-------UUAUU--------GCACUUUAAACAUUGUGGCCAUUUAAUAUUUCA .((......))...(((((((....((((((((((((((..........))))))))((.(.....-------...).--------))........)))))).)))))))......... ( -29.40) >DroEre_CAF1 190889 103 - 1 AUCCGAGCAGCAAAAGGUGGCAGGAACAGUGGUGCGGCAACGAUAUUAAUGUUAUACACUUGAAAU-------UUAAU--------GCAGUUUAAACAUUGCGGCUAUUUAAUAUU-AA .....(((.((((.(((((.....((((.(((((((....)).))))).))))...)))))....(-------((((.--------.....)))))..)))).)))..........-.. ( -21.60) >DroYak_CAF1 188877 117 - 1 AUCACAGCAGCAAAAAGUGCCACGAACAGUGGUGUGGCAACGAUGUUAAUGCCACACACUUAAAAUCUGUGACUUAACGAAAACUUUCAAUCUGAAAACUGCUGUACUAAAAUGUU-C- ...(((((((....(((((((((.....)))))((((((..........)))))).(((.........))))))).........((((.....)))).)))))))...........-.- ( -30.10) >consensus AGCCAAGCAGCAAAAAGUGGCAGGAACAGUGGUGUGGCAACGAUAUUAAUGCCACACGCUGGAAAU_______UUAAU________GCACUUUAAACAUUGUGGCCAUUUAAUAUUUCA ..............(((((((....((((((((((((((..........))))))))................(((................))).)))))).)))))))......... (-15.91 = -16.75 + 0.84)

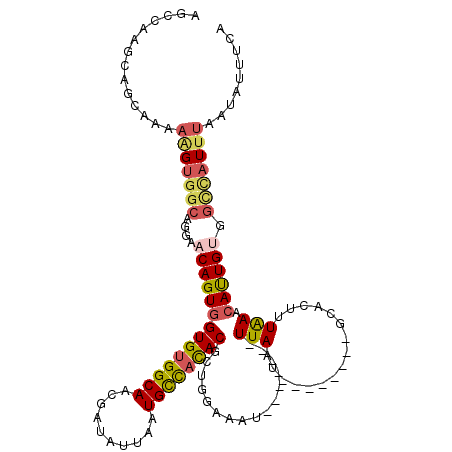
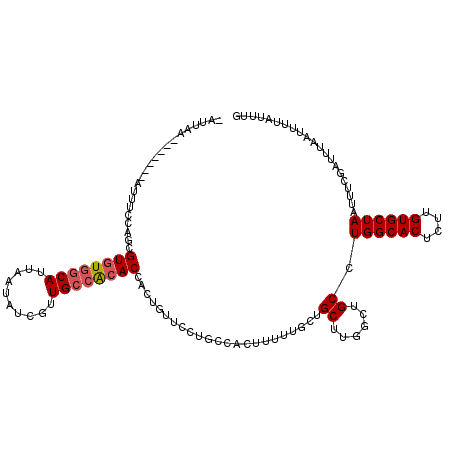
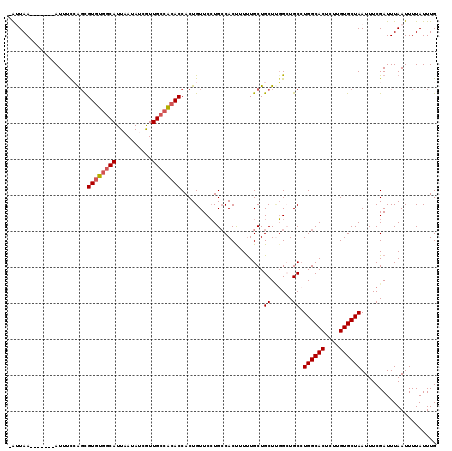
| Location | 4,753,872 – 4,753,983 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.69 |
| Mean single sequence MFE | -28.96 |
| Consensus MFE | -18.46 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4753872 111 + 22407834 -AAUAA-------AUUUUCAUCGUGUGGCAUUAAUACCAUUGCCACACCACUGUUCCUGCCACUUUCUGCUGCUUGGCUGCCUGGCACUCUUGUGCUAAUUUCGAUUUAAUUUUAUUUG -(((((-------(.((..(((((((((((..........))))))))..........((((............))))....((((((....)))))).....)))..)).)))))).. ( -27.20) >DroSec_CAF1 182339 111 + 1 -AUUAA-------AUUCCCGGCGUGUGGCAUUUAUAUCGUUGCCACACCACUGUUCCUGCCACUUUUUGCUGCUUGGCUGCAUGGCACUCUUGUGCUAAUUUCGAUUUAAUUUUAUUUG -(((((-------(((....((((((((((..........))))))))..........((((............)))).)).((((((....)))))).....))))))))........ ( -29.00) >DroSim_CAF1 178269 111 + 1 -AAUAA-------AUUUCCGGCGUGUGGCAUUAAAAUCGUUGCCACACCACUGUUCCUGCCACUUUUUGCUGCUUGGCUGCCUGGCACUCUUGUGCUAAUUUCGAUUUAAUUUUAUUUG -..(((-------(((...(((((((((((..........))))))))..........((((............)))).)))((((((....)))))).....)))))).......... ( -30.90) >DroEre_CAF1 190921 111 + 1 -AUUAA-------AUUUCAAGUGUAUAACAUUAAUAUCGUUGCCGCACCACUGUUCCUGCCACCUUUUGCUGCUCGGAUGCCUGGCACUCUGGUGCUAAUUUCGAUUUAAUUUUAUUUG -.....-------...........((((.(((((.((((.....((((((..((.((.((..((...........))..))..)).))..))))))......))))))))).))))... ( -21.00) >DroYak_CAF1 188915 119 + 1 CGUUAAGUCACAGAUUUUAAGUGUGUGGCAUUAACAUCGUUGCCACACCACUGUUCGUGGCACUUUUUGCUGCUGUGAUGCCUGGCACUCUGGUGCUAAUUUCGAUUUAAUUUUAUUUG .((((((((.(((......(((((((((((..........))))))).))))..(((..((((.....).)))..)))...)))((((....)))).......))))))))........ ( -36.70) >consensus _AUUAA_______AUUUCCAGCGUGUGGCAUUAAUAUCGUUGCCACACCACUGUUCCUGCCACUUUUUGCUGCUUGGCUGCCUGGCACUCUUGUGCUAAUUUCGAUUUAAUUUUAUUUG ......................((((((((..........)))))))).......................((......)).((((((....))))))..................... (-18.46 = -18.90 + 0.44)

| Location | 4,753,872 – 4,753,983 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.69 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -17.80 |
| Energy contribution | -17.72 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
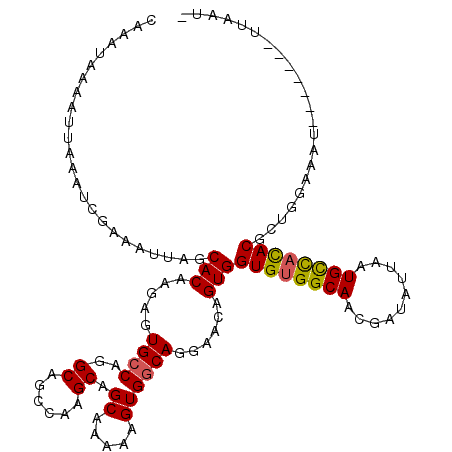
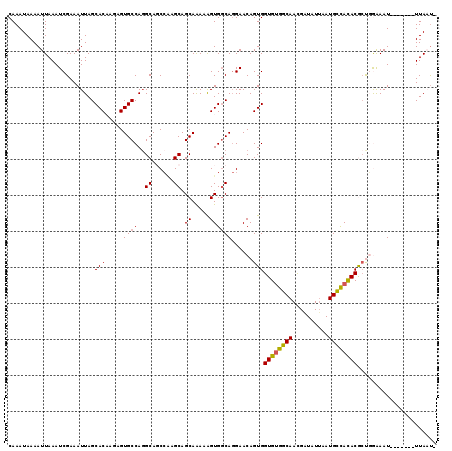
>2L_DroMel_CAF1 4753872 111 - 22407834 CAAAUAAAAUUAAAUCGAAAUUAGCACAAGAGUGCCAGGCAGCCAAGCAGCAGAAAGUGGCAGGAACAGUGGUGUGGCAAUGGUAUUAAUGCCACACGAUGAAAAU-------UUAUU- ..((((((.((..(((.......((((....))))......((((..(....)....))))..........((((((((..........)))))))))))..)).)-------)))))- ( -25.50) >DroSec_CAF1 182339 111 - 1 CAAAUAAAAUUAAAUCGAAAUUAGCACAAGAGUGCCAUGCAGCCAAGCAGCAAAAAGUGGCAGGAACAGUGGUGUGGCAACGAUAUAAAUGCCACACGCCGGGAAU-------UUAAU- ......(((((...(((......((((....))))...((.((((............))))..........((((((((..........))))))))))))).)))-------))...- ( -26.50) >DroSim_CAF1 178269 111 - 1 CAAAUAAAAUUAAAUCGAAAUUAGCACAAGAGUGCCAGGCAGCCAAGCAGCAAAAAGUGGCAGGAACAGUGGUGUGGCAACGAUUUUAAUGCCACACGCCGGAAAU-------UUAUU- ..((((((...............((((....))))..(((.((((............))))..........((((((((..........))))))))))).....)-------)))))- ( -28.70) >DroEre_CAF1 190921 111 - 1 CAAAUAAAAUUAAAUCGAAAUUAGCACCAGAGUGCCAGGCAUCCGAGCAGCAAAAGGUGGCAGGAACAGUGGUGCGGCAACGAUAUUAAUGUUAUACACUUGAAAU-------UUAAU- ...((((.((((((((.......((((((..((.((.(.((.((...........)))).).)).))..))))))(....)))).))))).))))...........-------.....- ( -24.10) >DroYak_CAF1 188915 119 - 1 CAAAUAAAAUUAAAUCGAAAUUAGCACCAGAGUGCCAGGCAUCACAGCAGCAAAAAGUGCCACGAACAGUGGUGUGGCAACGAUGUUAAUGCCACACACUUAAAAUCUGUGACUUAACG .......................((((....))))......((((((.......(((((((((.....))))(((((((..........)))))))))))).....))))))....... ( -28.70) >consensus CAAAUAAAAUUAAAUCGAAAUUAGCACAAGAGUGCCAGGCAGCCAAGCAGCAAAAAGUGGCAGGAACAGUGGUGUGGCAACGAUAUUAAUGCCACACGCUGGAAAU_______UUAAU_ ........................(((.....((((..((......)).((.....))))))......)))((((((((..........))))))))...................... (-17.80 = -17.72 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:44 2006