| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,750,411 – 4,750,505 |
| Length | 94 |
| Max. P | 0.883010 |

| Location | 4,750,411 – 4,750,505 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.65 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -18.93 |
| Energy contribution | -19.46 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4750411 94 + 22407834 ------------GUGGCAAGAGUAAAAGCAAUUUAGUUUACUGCAAAUGCUGG-CCGGCGGGCCUUUGUUCUGUUC----------UGCUGCCUUCAGCAUUUUGGCUGAUUCAUGG ------------(..(((((((((((((((.(((((....))).)).))))((-((....))))))))))))....----------)))..)..(((((......)))))....... ( -26.20) >DroPse_CAF1 216621 102 + 1 GCAUAAGCAGUAGCAGCAGCAGUAAAAGCAAUUUAGUUUACUGCAAAUGUUAG-CCGGCGGC----UACAAUGUUC----------UGCUGCCCUCAGCAUUUUAGCUGAUUCAUGG .(((..(((((((.(((((((((((..((......)))))))))...(((.((-((...)))----)))).)))))----------))))))..(((((......)))))...))). ( -35.10) >DroEre_CAF1 187412 94 + 1 ------------GUGGCAAGAGUAAAAGCAAUUUAGUUUACUGCAAAUGCUGG-CCGGCGGCUCUUUGUUCUGUUC----------UGCUGCCUUCAGCAUUUUGGCUGAUUCAUGG ------------....((.((((...(((....(((....))).(((((((((-..((((((..............----------.)))))).)))))))))..))).)))).)). ( -26.86) >DroYak_CAF1 185448 104 + 1 ------------GUGGCAAGAGUAAAAGCAAUUUAGUUUACUGCAAAUGCUGG-CCGGCGGCCCUUUGUUCUGCUCUGUACUGUUGUGCUGCCUUCAGCAUUUUGGCUGAUUCAUGG ------------..((((((((((..((((.(((((....))).)).))))((-((...))))........))))))((((....)))))))).(((((......)))))....... ( -31.10) >DroAna_CAF1 198023 95 + 1 ------------GAGGCGGCAGUAAAAGCAAUUUAGUUUACUGCAAAUGUUGGGCUGGCCUGGCGGUGCAUUGUUC----------UGCUGCCUUCAGCAUUUUAGCUGAUUCAUGG ------------.(((((((((...((((......))))..((((..(((..((....))..))).)))).....)----------))))))))(((((......)))))....... ( -35.10) >DroPer_CAF1 224337 102 + 1 GCAUAAGCAGUAGCAGCAGCAGUAAAAGCAAUUUAGUUUACUGCAAAUGUUAG-CCGGCGGC----UACAAUGUUC----------UGCUGCCCUCAGCAUUUUAGCUGAUUCAUGG .(((..(((((((.(((((((((((..((......)))))))))...(((.((-((...)))----)))).)))))----------))))))..(((((......)))))...))). ( -35.10) >consensus ____________GUGGCAACAGUAAAAGCAAUUUAGUUUACUGCAAAUGCUGG_CCGGCGGC_CUUUGCACUGUUC__________UGCUGCCUUCAGCAUUUUAGCUGAUUCAUGG .............((((...(((((..((......)))))))..(((((((((...((((((.........................)))))).)))))))))..))))........ (-18.93 = -19.46 + 0.53)

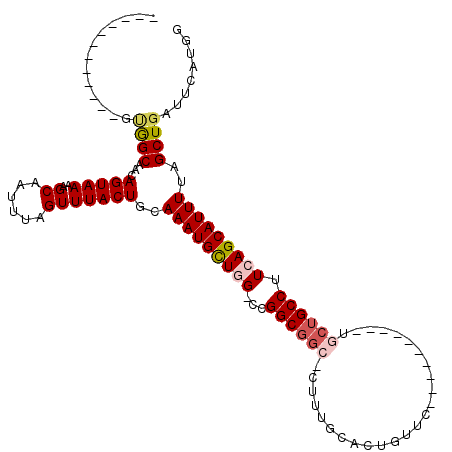
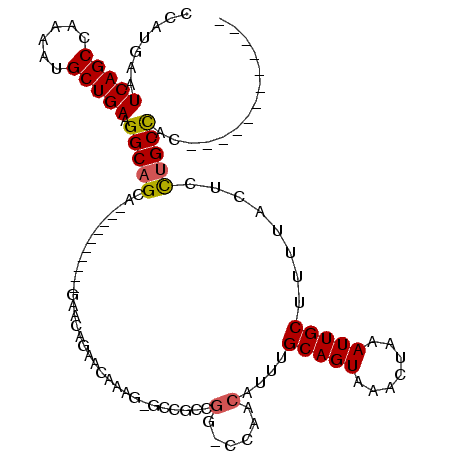
| Location | 4,750,411 – 4,750,505 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.65 |
| Mean single sequence MFE | -27.76 |
| Consensus MFE | -13.90 |
| Energy contribution | -13.77 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.883010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4750411 94 - 22407834 CCAUGAAUCAGCCAAAAUGCUGAAGGCAGCA----------GAACAGAACAAAGGCCCGCCGG-CCAGCAUUUGCAGUAAACUAAAUUGCUUUUACUCUUGCCAC------------ .......(((((......))))).(((((.(----------(...........((((....))-))((((.((..((....)).)).))))....)).)))))..------------ ( -25.40) >DroPse_CAF1 216621 102 - 1 CCAUGAAUCAGCUAAAAUGCUGAGGGCAGCA----------GAACAUUGUA----GCCGCCGG-CUAACAUUUGCAGUAAACUAAAUUGCUUUUACUGCUGCUGCUACUGCUUAUGC .(((((.(((((......))))).(((((((----------......((((----(((...))-)).)))...((((((((..........))))))))))))))).....))))). ( -33.80) >DroEre_CAF1 187412 94 - 1 CCAUGAAUCAGCCAAAAUGCUGAAGGCAGCA----------GAACAGAACAAAGAGCCGCCGG-CCAGCAUUUGCAGUAAACUAAAUUGCUUUUACUCUUGCCAC------------ .......(((((......))))).(((.((.----------..............)).)))((-(.((.....(((((.......))))).......)).)))..------------ ( -20.96) >DroYak_CAF1 185448 104 - 1 CCAUGAAUCAGCCAAAAUGCUGAAGGCAGCACAACAGUACAGAGCAGAACAAAGGGCCGCCGG-CCAGCAUUUGCAGUAAACUAAAUUGCUUUUACUCUUGCCAC------------ .......(((((......))))).(((((......((((.(((((((.......((((...))-)).((....))...........))))))))))).)))))..------------ ( -27.50) >DroAna_CAF1 198023 95 - 1 CCAUGAAUCAGCUAAAAUGCUGAAGGCAGCA----------GAACAAUGCACCGCCAGGCCAGCCCAACAUUUGCAGUAAACUAAAUUGCUUUUACUGCCGCCUC------------ .......(((((......)))))((((.(((----------(.....((((......((....)).......))))((((......)))).....)))).)))).------------ ( -25.12) >DroPer_CAF1 224337 102 - 1 CCAUGAAUCAGCUAAAAUGCUGAGGGCAGCA----------GAACAUUGUA----GCCGCCGG-CUAACAUUUGCAGUAAACUAAAUUGCUUUUACUGCUGCUGCUACUGCUUAUGC .(((((.(((((......))))).(((((((----------......((((----(((...))-)).)))...((((((((..........))))))))))))))).....))))). ( -33.80) >consensus CCAUGAAUCAGCCAAAAUGCUGAAGGCAGCA__________GAACAGAACAAAG_GCCGCCGG_CCAACAUUUGCAGUAAACUAAAUUGCUUUUACUCCUGCCAC____________ .......(((((......))))).(((((................................(......)....(((((.......)))))........))))).............. (-13.90 = -13.77 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:38 2006