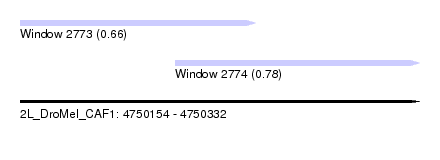
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,750,154 – 4,750,332 |
| Length | 178 |
| Max. P | 0.777295 |

| Location | 4,750,154 – 4,750,259 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 88.16 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -20.74 |
| Energy contribution | -20.30 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4750154 105 + 22407834 CGUACGCAAAAUGUUUGAGCUGGCAGUUGCCAAAGCGACGGGAACUGAAACAGUAACUAGCACAACUAGAGCAAAACUAGCAAAACCAGCACAACCGACACAAGU ...........((.(((.((((((((((.((........)).))))).........((((.....)))).((.......))....))))).))).))........ ( -22.60) >DroEre_CAF1 187134 94 + 1 CGUACGCAAAAUGUUUGAGCUGGCAGUUGCCAAGGCGACGGGAACUGAAACAGCAACUAGCACAACUU-----------GCACAACCAGCACAACCGACACAAGU ...........((.(((.((((((((((.((........)).)))))............(((.....)-----------))....))))).))).))........ ( -21.50) >DroYak_CAF1 185194 94 + 1 AGUACGCAAAAUGUUUGAGCUGGCAGUUGCCAAAGCGACGGGAACUGAAAUAGCAACUAGCACAACUU-----------GCACAACCAGCACAACCAACACAAGU ...........((.(((.((((((((((.((........)).)))))............(((.....)-----------))....))))).))).))........ ( -22.20) >consensus CGUACGCAAAAUGUUUGAGCUGGCAGUUGCCAAAGCGACGGGAACUGAAACAGCAACUAGCACAACUU___________GCACAACCAGCACAACCGACACAAGU ...........((.(((.((((((((((.((........)).))))).....((.....))........................))))).))).))........ (-20.74 = -20.30 + -0.44)



| Location | 4,750,223 – 4,750,332 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 88.41 |
| Mean single sequence MFE | -22.72 |
| Consensus MFE | -17.54 |
| Energy contribution | -17.18 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4750223 109 + 22407834 ----AGCAAAACUAGCAAAACCAGCACAACCGACACAAGUGCAACUCCGACUGCAGGACAGCACUCAUACACGCGCGCACACGGUGCUCUAAGUGCUUUUUGACAUGCAACUA ----.(((.......(((((..(((((..........(((((...(((.......)))..))))).........((((.....)))).....))))))))))...)))..... ( -24.70) >DroSec_CAF1 178814 113 + 1 AACUAGCAAAACUAGCAAAACCAGCACAACCGACACAAGUGCAACUCCGACUGCAGGACAGCACCCAUACACGCGCACACACGGCGCUUUAAGUGCUUUUUGACAUGCAACUA .....(((.......(((((..(((((...........((((...(((.......)))..))))........((((.......)))).....))))))))))...)))..... ( -25.30) >DroSim_CAF1 174589 104 + 1 A---------ACUAGCAAAACCAGCACAACCGACACAAGUGCAACUCCGACUGCAGGACAGCACCCAUACACGCGCACACACGGCGCUCUAAGUGCUUUUUGACAUGCAACUA .---------....((((((..(((((...........((((...(((.......)))..))))........((((.......)))).....)))))))))).)......... ( -23.50) >DroEre_CAF1 187202 99 + 1 --------------GCACAACCAGCACAACCGACACAAGUGCAACUCCGACUGCAGGACAGCACCUAUACACUCGCACAUACGGAGCUCUAAGUGCUUUUUGACAUGCAACUA --------------((((((..(((((...........((((...(((.......)))..)))).......(((.(......))))......)))))..)))...)))..... ( -19.80) >DroYak_CAF1 185262 99 + 1 --------------GCACAACCAGCACAACCAACACAAGUGCAACUCCGACUGCAGGACAGCACCCAUACACUCGCACACACGCAGCUCUAAGUGCUUUUUGACAUGCAACUA --------------((((((..(((((...........((((...(((.......)))..))))..........((......))........)))))..)))...)))..... ( -20.30) >consensus __________ACUAGCAAAACCAGCACAACCGACACAAGUGCAACUCCGACUGCAGGACAGCACCCAUACACGCGCACACACGGAGCUCUAAGUGCUUUUUGACAUGCAACUA ..............(((......((((...........)))).........(((((((.(((((.......(((.(......))))......)))))))))).)))))..... (-17.54 = -17.18 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:37 2006