
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,748,603 – 4,748,714 |
| Length | 111 |
| Max. P | 0.997513 |

| Location | 4,748,603 – 4,748,714 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -24.16 |
| Energy contribution | -26.46 |
| Covariance contribution | 2.30 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
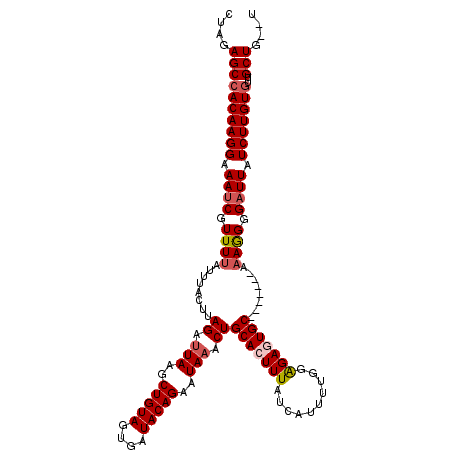
>2L_DroMel_CAF1 4748603 111 + 22407834 CUCGAGCCACAAGGAAAACGUUUUAUUUACUUAGAUUAAGCUGUAGUGAUACAGAAUAAACUGCACUUUAUCAUUUUGGAGAGUGC-------AAAAGGGAUUAUCUUGUGUUGCUG--U ....((((((((((((..(.((((........((.(((..(((((....)))))..))).))(((((((.((.....)))))))))-------)))).)..)).)))))))..))).--. ( -28.30) >DroSec_CAF1 177210 111 + 1 CUGGAGCCACAAGGAAAUCGUUUUAUUUACUUAGAUUAAGCUGUAGUGAUACAGAAUAAACUGCACUUUAUCAUUUUGGAGAGUGC-------AAAGGGGAUUAUCUUGUGUUGCUG--U ....((((((((((.((((.((((...........(((..(((((....)))))..)))..((((((((.((.....)))))))))-------))))).)))).)))))))..))).--. ( -32.00) >DroSim_CAF1 172965 111 + 1 CUGGAGCCACAAGGAAAUCGUUUUAUUUACUUAGAUUAAGCUGUAGUGAUACAGAAUAAACUGCACUUUAUCAUUUUGGAGAGUGC-------AAAGGGGAUUAUCUUGUGUUGCUG--U ....((((((((((.((((.((((...........(((..(((((....)))))..)))..((((((((.((.....)))))))))-------))))).)))).)))))))..))).--. ( -32.00) >DroEre_CAF1 185557 112 + 1 CUAGAGCCACAAGGAAAUCGUUUUAUUUACUUAGACUAAGCUGUAGUGAUACAGAAUAAACUGCACUUUAUCAUUUUAGGGAAUGCA--------AGGGGAUUAUCUUGUGUUGCUGUGU .((.((((((((((.((((.((((................(((((....)))))........((((((((......)))))..))))--------))).)))).)))))))..))).)). ( -26.90) >DroYak_CAF1 183612 118 + 1 CUAGAGCCACAAGGAAAUCGUUUUAUUUACUUAGAUUAAGCUGUAGUGAUACAGAAUAAACUGCACUUUAUCAUUUUGCAGAAUGCAGAAUGCAAAAGGGAUUAUCUUGUAUUGCUG--U ....(((.((((((.((((............(((.(((..(((((....)))))..))).)))..((((..((((((((.....))))))))..)))).)))).))))))...))).--. ( -30.60) >consensus CUAGAGCCACAAGGAAAUCGUUUUAUUUACUUAGAUUAAGCUGUAGUGAUACAGAAUAAACUGCACUUUAUCAUUUUGGAGAGUGC_______AAAGGGGAUUAUCUUGUGUUGCUG__U ....((((((((((.((((.((((........((.(((..(((((....)))))..))).))(((((((..........)))))))........)))).)))).)))))))..))).... (-24.16 = -26.46 + 2.30)

| Location | 4,748,603 – 4,748,714 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -21.41 |
| Consensus MFE | -20.48 |
| Energy contribution | -20.12 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4748603 111 - 22407834 A--CAGCAACACAAGAUAAUCCCUUUU-------GCACUCUCCAAAAUGAUAAAGUGCAGUUUAUUCUGUAUCACUACAGCUUAAUCUAAGUAAAUAAAACGUUUUCCUUGUGGCUCGAG .--.(((..((((((............-------(((((.((......))...)))))((.(((..(((((....)))))..))).))...................))))))))).... ( -19.40) >DroSec_CAF1 177210 111 - 1 A--CAGCAACACAAGAUAAUCCCCUUU-------GCACUCUCCAAAAUGAUAAAGUGCAGUUUAUUCUGUAUCACUACAGCUUAAUCUAAGUAAAUAAAACGAUUUCCUUGUGGCUCCAG .--.(((..((((((..((((......-------(((((.((......))...)))))((.(((..(((((....)))))..))).)).............))))..))))))))).... ( -22.50) >DroSim_CAF1 172965 111 - 1 A--CAGCAACACAAGAUAAUCCCCUUU-------GCACUCUCCAAAAUGAUAAAGUGCAGUUUAUUCUGUAUCACUACAGCUUAAUCUAAGUAAAUAAAACGAUUUCCUUGUGGCUCCAG .--.(((..((((((..((((......-------(((((.((......))...)))))((.(((..(((((....)))))..))).)).............))))..))))))))).... ( -22.50) >DroEre_CAF1 185557 112 - 1 ACACAGCAACACAAGAUAAUCCCCU--------UGCAUUCCCUAAAAUGAUAAAGUGCAGUUUAUUCUGUAUCACUACAGCUUAGUCUAAGUAAAUAAAACGAUUUCCUUGUGGCUCUAG ....(((..((((((..((((....--------.(((((..............)))))((.(((..(((((....)))))..))).)).............))))..))))))))).... ( -19.44) >DroYak_CAF1 183612 118 - 1 A--CAGCAAUACAAGAUAAUCCCUUUUGCAUUCUGCAUUCUGCAAAAUGAUAAAGUGCAGUUUAUUCUGUAUCACUACAGCUUAAUCUAAGUAAAUAAAACGAUUUCCUUGUGGCUCUAG .--.(((..((((((..((((...(((((...(((((((.(((.....).)).))))))).(((..(((((....)))))..))).....)))))......))))..))))))))).... ( -23.20) >consensus A__CAGCAACACAAGAUAAUCCCCUUU_______GCACUCUCCAAAAUGAUAAAGUGCAGUUUAUUCUGUAUCACUACAGCUUAAUCUAAGUAAAUAAAACGAUUUCCUUGUGGCUCCAG ....(((..((((((..((((.............(((((..............)))))((.(((..(((((....)))))..))).)).............))))..))))))))).... (-20.48 = -20.12 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:34 2006