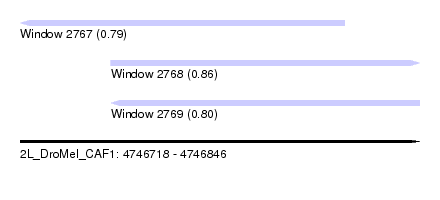
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,746,718 – 4,746,846 |
| Length | 128 |
| Max. P | 0.859672 |

| Location | 4,746,718 – 4,746,822 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.98 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -19.13 |
| Energy contribution | -19.63 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4746718 104 - 22407834 AAAGGAAAGCG-----UGGCAAACAUGCAUGACAAUGCAUGACUAAAGUAAUGAAUUACUUAAGUCCCUCGCCCACUGCAAC-----CAACAAGACAUAUGCCC------CCCAUUCCGA ...((((.(.(-----.((((...((((((....))))))((((.((((((....)))))).))))....((.....))...-----............)))).------).).)))).. ( -25.40) >DroPse_CAF1 212125 106 - 1 -AAAGAAAGCG-----UGGCAAACAUGCAUGACAAUGCAUGACAAAAGUAAUGAAUUACUUAAGUCCCUCGCCCACUGCACA---CACAACAAGACAUAUGGCC-----CCCGAUCCGGG -.......(((-----((((...(((((((....)))))))....((((((....)))))).........).)))).))...---...................-----((((...)))) ( -24.00) >DroEre_CAF1 183678 105 - 1 AAAGGAAAGCG-----UGGCAAACAUGCAUGACAAUGCAUGACUAAAGUAAUGAAUUACUUAAGUCCCUCGCCCACUGCAAC-----CAACAGCACAUAUGUCC-----CCCAAUUCCGA ...((((.(((-----.((.....((((((....))))))((((.((((((....)))))).)))))).)))....(((...-----.....))).........-----.....)))).. ( -26.50) >DroYak_CAF1 181704 106 - 1 AAAGGAAAGCGUGGCGUGGCAAACAUGCAUGACAAUGCAUGACUAAAGUAAUGAAUUACUUAAGUCCCUCGCCCACUGCAGC-----CCCC----CAUUCCGAA-----ACCGAAUCCGA ...(((..((((((((.((.....((((((....))))))((((.((((((....)))))).)))))).)).)))).))...-----....----.....((..-----..))..))).. ( -30.50) >DroAna_CAF1 194502 114 - 1 AUACCAAAGCG-----UGGCAAACAUGCAUGACAAUGCAUGACUAAAGUAAUGAAUUACUUAAG-CCCUCGCCCACUGCACUGAACACAGCAAGACAUAUGGCCAAAGAACCGGAUCAGA ...(((..(((-----.(((...(((((((....)))))))....((((((....))))))..)-))..)))...((...(((....)))..)).....)))((........))...... ( -25.80) >DroPer_CAF1 219827 106 - 1 -AAAGAAAGCG-----UGGCAAACAUGCAUGACAAUGCAUGACAAAAGUAAUGAAUUACUUAAGUCCCUCGCCCACUGCACA---CACAACAAGACAUAUGGCC-----CCCGAUCCGGG -.......(((-----((((...(((((((....)))))))....((((((....)))))).........).)))).))...---...................-----((((...)))) ( -24.00) >consensus AAAGGAAAGCG_____UGGCAAACAUGCAUGACAAUGCAUGACUAAAGUAAUGAAUUACUUAAGUCCCUCGCCCACUGCACC_____CAACAAGACAUAUGGCC_____CCCGAUUCCGA ........(((......(((...(((((((....)))))))(((.((((((....)))))).))).....)))...)))......................................... (-19.13 = -19.63 + 0.50)



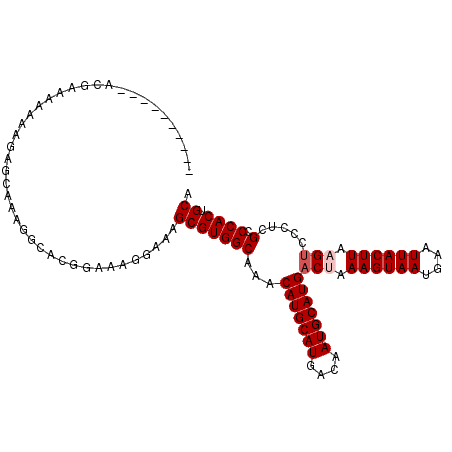
| Location | 4,746,747 – 4,746,846 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.94 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -22.87 |
| Energy contribution | -23.37 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4746747 99 + 22407834 UGCAGUGGGCGAGGGACUUAAGUAAUUCAUUACUUUAGUCAUGCAUUGUCAUGCAUGUUUGCCACGCUUUCCUUUCCGUGCCUUUGCGAUCCUUUCCGU---------- .((((.(((((.((((((.((((((....)))))).))))((((((....))))))...................)).)))))))))............---------- ( -27.20) >DroPse_CAF1 212157 99 + 1 UGCAGUGGGCGAGGGACUUAAGUAAUUCAUUACUUUUGUCAUGCAUUGUCAUGCAUGUUUGCCACGCUUUCUUU--CAUUCCUUCCCUCUUUUUUUCGU--------GU ...((((((((((.(((..((((((....))))))..)))((((((....)))))).)))))).))))......--.......................--------.. ( -24.20) >DroSec_CAF1 175442 99 + 1 UGCAGUGGGCGAGGGACUUAAGUAAUUCAUUACUUUAGUCAUGCAUUGUCAUGCAUGUUUGCCACGCUUUCCUUUCCGUGCCUUUGCAAUUUUUUCCGC---------- (((((.(((((.((((((.((((((....)))))).))))((((((....))))))...................)).))))))))))...........---------- ( -27.90) >DroSim_CAF1 171140 99 + 1 UGCAGUGGGCGAGGGACUUAAGUAAUUCAUUACUUUAGUCAUGCAUUGUCAUGCAUGUUUGCCACGCUUUCCUUUCCGUGCCUUUGCCAUCCUUUCCGC---------- .......(((((((((((.((((((....)))))).)))(((((((....))))))).....((((..........))))))))))))...........---------- ( -26.70) >DroAna_CAF1 194542 108 + 1 UGCAGUGGGCGAGGG-CUUAAGUAAUUCAUUACUUUAGUCAUGCAUUGUCAUGCAUGUUUGCCACGCUUUGGUAUCCUUGCCGCUUCCCUCCAGUUCCAGGCUCUGACU ..(((.(((((((((-((.((((((....)))))).)))(((((((....)))))))..(((((.....))))).))))))).....(((........)))).)))... ( -32.00) >DroPer_CAF1 219859 99 + 1 UGCAGUGGGCGAGGGACUUAAGUAAUUCAUUACUUUUGUCAUGCAUUGUCAUGCAUGUUUGCCACGCUUUCUUU--CAUUCCUUCCCUCUUUUUUUCGU--------GU ...((((((((((.(((..((((((....))))))..)))((((((....)))))).)))))).))))......--.......................--------.. ( -24.20) >consensus UGCAGUGGGCGAGGGACUUAAGUAAUUCAUUACUUUAGUCAUGCAUUGUCAUGCAUGUUUGCCACGCUUUCCUUUCCGUGCCUUUGCCAUCCUUUCCGU__________ ...((((((((((.((((.((((((....)))))).))))((((((....)))))).)))))).))))......................................... (-22.87 = -23.37 + 0.50)

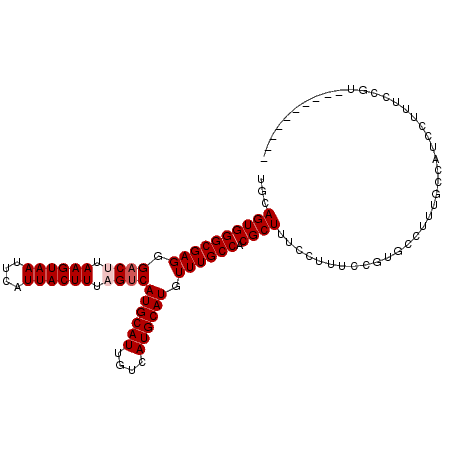
| Location | 4,746,747 – 4,746,846 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.94 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -20.85 |
| Energy contribution | -21.35 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4746747 99 - 22407834 ----------ACGGAAAGGAUCGCAAAGGCACGGAAAGGAAAGCGUGGCAAACAUGCAUGACAAUGCAUGACUAAAGUAAUGAAUUACUUAAGUCCCUCGCCCACUGCA ----------..((..(((........(.((((..........)))).)...(((((((....)))))))(((.((((((....)))))).))).)))..))....... ( -25.50) >DroPse_CAF1 212157 99 - 1 AC--------ACGAAAAAAAGAGGGAAGGAAUG--AAAGAAAGCGUGGCAAACAUGCAUGACAAUGCAUGACAAAAGUAAUGAAUUACUUAAGUCCCUCGCCCACUGCA ..--------..........((((((.......--.................(((((((....)))))))....((((((....))))))...))))))((.....)). ( -23.80) >DroSec_CAF1 175442 99 - 1 ----------GCGGAAAAAAUUGCAAAGGCACGGAAAGGAAAGCGUGGCAAACAUGCAUGACAAUGCAUGACUAAAGUAAUGAAUUACUUAAGUCCCUCGCCCACUGCA ----------((((.......(((....)))......((...(((.((.....((((((....))))))((((.((((((....)))))).)))))).))))).)))). ( -28.60) >DroSim_CAF1 171140 99 - 1 ----------GCGGAAAGGAUGGCAAAGGCACGGAAAGGAAAGCGUGGCAAACAUGCAUGACAAUGCAUGACUAAAGUAAUGAAUUACUUAAGUCCCUCGCCCACUGCA ----------((((........((....)).......((...(((.((.....((((((....))))))((((.((((((....)))))).)))))).))))).)))). ( -28.00) >DroAna_CAF1 194542 108 - 1 AGUCAGAGCCUGGAACUGGAGGGAAGCGGCAAGGAUACCAAAGCGUGGCAAACAUGCAUGACAAUGCAUGACUAAAGUAAUGAAUUACUUAAG-CCCUCGCCCACUGCA ........(((........)))...((((...((...))...(((.(((...(((((((....)))))))....((((((....))))))..)-))..)))...)))). ( -28.10) >DroPer_CAF1 219859 99 - 1 AC--------ACGAAAAAAAGAGGGAAGGAAUG--AAAGAAAGCGUGGCAAACAUGCAUGACAAUGCAUGACAAAAGUAAUGAAUUACUUAAGUCCCUCGCCCACUGCA ..--------..........((((((.......--.................(((((((....)))))))....((((((....))))))...))))))((.....)). ( -23.80) >consensus __________ACGAAAAAAAGAGCAAAGGCACGGAAAGGAAAGCGUGGCAAACAUGCAUGACAAUGCAUGACUAAAGUAAUGAAUUACUUAAGUCCCUCGCCCACUGCA ..........................................(((((((...(((((((....)))))))(((.((((((....)))))).))).....).)))).)). (-20.85 = -21.35 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:32 2006