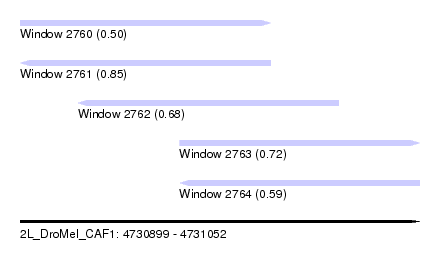
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,730,899 – 4,731,052 |
| Length | 153 |
| Max. P | 0.851003 |

| Location | 4,730,899 – 4,730,995 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.68 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -18.55 |
| Energy contribution | -19.05 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4730899 96 + 22407834 -------AGCCG-----------GGGGGCGUGGCAACGUCUGUUGUGUGUUAUUACCACCGCAGUCAUUAAAUCUU-UUGUGUGUAAUUAAAAAAUUACAAUUUAAUGCUUUGCG----- -------.((((-----------(.((((((....)))))).))).))...........(((((.((((((((...-.....(((((((....)))))))))))))))..)))))----- ( -24.00) >DroPse_CAF1 187625 102 + 1 -------GGCCA-GGGCACU----GGGGCGUGGCAACGUCUGUUGUGUGUUAUUACCACCGCAGUCAUUAAAUCUU-UUGUGUGUAAUUAAAAAAUUACAAUUUAAUGCCUGGCG----- -------.((((-((((((.----.(((...(....)(.((((.(((.((....))))).)))).).......)))-..))))((((((....)))))).........)))))).----- ( -32.00) >DroEre_CAF1 167359 100 + 1 GC-----GCCC---------GGAGGGGGCGUGGCAGCGUCUGUUGUGUGUUAUUACCACCGCAGUCAUUAAAUCUU-UUGUGUGUAAUUAAAAAAUUACAAUUUAAUGCUUUGCG----- ((-----((((---------.....)))))).((((((.((((.(((.((....))))).)))).)(((((((...-.....(((((((....))))))))))))))))..))).----- ( -29.40) >DroWil_CAF1 166312 109 + 1 -------GGCCA-ACGUAUC---GAGGGAGAAAUAACGUCUGUUGUGUGUUAUUACCACCGCAGUCAUUAAAUCUUUUUGUGUGUAAUUAAAAAAUUACAAUUUAAUGCUUGGCAAAUAG -------.((((-(.((((.---.(((((((..(((.(.((((.(((.((....))))).)))).).)))..))))).....(((((((....))))))).))..)))))))))...... ( -27.40) >DroYak_CAF1 165821 109 + 1 GC-----GGCCUGGGGCGGUGGGGUGGGCGUGGCAGCGUCUGUUGUGUGUUAUUACCACCGCAGUCAUUAAAUCUU-UUGUGUGUAAUUAAAAAAUUACAAUUUAAUGCUUUGCG----- ((-----((......(((((((...(.(((..((((...))))..))).).....)))))))((.((((((((...-.....(((((((....))))))))))))))))))))).----- ( -34.20) >DroAna_CAF1 178627 102 + 1 -CCAUGAGACUA-----------GGGGGCGUUGCAGCGUCUGUUGUGUGUUAUUACCACCACAGUCAUUAAAUAUU-UUGUACGUAAUUAAAAAAUUACAAUUUAAUGCUUUGCG----- -...........-----------....(((..(((..(.((((.(((.((....))))).)))).)..........-......((((((....)))))).......)))..))).----- ( -20.40) >consensus _______GGCCA___________GGGGGCGUGGCAACGUCUGUUGUGUGUUAUUACCACCGCAGUCAUUAAAUCUU_UUGUGUGUAAUUAAAAAAUUACAAUUUAAUGCUUUGCG_____ ...........................(((.((((..(.((((.(((.((....))))).)))).)................(((((((....)))))))......)))).)))...... (-18.55 = -19.05 + 0.50)


| Location | 4,730,899 – 4,730,995 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.68 |
| Mean single sequence MFE | -24.69 |
| Consensus MFE | -15.90 |
| Energy contribution | -15.77 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4730899 96 - 22407834 -----CGCAAAGCAUUAAAUUGUAAUUUUUUAAUUACACACAA-AAGAUUUAAUGACUGCGGUGGUAAUAACACACAACAGACGUUGCCACGCCCCC-----------CGGCU------- -----((((...(((((((((((((((....))))))).....-...))))))))..))))((((((((..(........)..))))))))(((...-----------.))).------- ( -26.00) >DroPse_CAF1 187625 102 - 1 -----CGCCAGGCAUUAAAUUGUAAUUUUUUAAUUACACACAA-AAGAUUUAAUGACUGCGGUGGUAAUAACACACAACAGACGUUGCCACGCCCC----AGUGCCC-UGGCC------- -----.(((((((((((((((((((((....))))))).....-...))))))))((((.(((((((((..(........)..))))))..))).)----)))..))-)))).------- ( -30.90) >DroEre_CAF1 167359 100 - 1 -----CGCAAAGCAUUAAAUUGUAAUUUUUUAAUUACACACAA-AAGAUUUAAUGACUGCGGUGGUAAUAACACACAACAGACGCUGCCACGCCCCCUCC---------GGGC-----GC -----((((...(((((((((((((((....))))))).....-...))))))))..))))((((((..................))))))((((.....---------))))-----.. ( -27.07) >DroWil_CAF1 166312 109 - 1 CUAUUUGCCAAGCAUUAAAUUGUAAUUUUUUAAUUACACACAAAAAGAUUUAAUGACUGCGGUGGUAAUAACACACAACAGACGUUAUUUCUCCCUC---GAUACGU-UGGCC------- ......(((((.........(((((((....))))))).......(((..(((((.(((..(((((....)).)))..))).)))))..))).....---......)-)))).------- ( -24.10) >DroYak_CAF1 165821 109 - 1 -----CGCAAAGCAUUAAAUUGUAAUUUUUUAAUUACACACAA-AAGAUUUAAUGACUGCGGUGGUAAUAACACACAACAGACGCUGCCACGCCCACCCCACCGCCCCAGGCC-----GC -----.((..(((((((((((((((((....))))))).....-...)))))))).))(((((((........(......)..((......)).....))))))).....)).-----.. ( -22.10) >DroAna_CAF1 178627 102 - 1 -----CGCAAAGCAUUAAAUUGUAAUUUUUUAAUUACGUACAA-AAUAUUUAAUGACUGUGGUGGUAAUAACACACAACAGACGCUGCAACGCCCCC-----------UAGUCUCAUGG- -----.(((...(((((((((((((((....))))))).....-...)))))))).((((.(((((....)).))).))))....))).........-----------...........- ( -18.00) >consensus _____CGCAAAGCAUUAAAUUGUAAUUUUUUAAUUACACACAA_AAGAUUUAAUGACUGCGGUGGUAAUAACACACAACAGACGCUGCCACGCCCCC___________UGGCC_______ .....((((...(((((((((((((((....))))))).........))))))))..))))((((((..................))))))............................. (-15.90 = -15.77 + -0.13)

| Location | 4,730,921 – 4,731,021 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.32 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -17.42 |
| Energy contribution | -17.67 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4730921 100 - 22407834 GACCGC-----ACGCCGUGUCAAUAAAUGA-----G---------CGCAAAGCAUUAAAUUGUAAUUUUUUAAUUACACACAA-AAGAUUUAAUGACUGCGGUGGUAAUAACACACAACA .(((((-----.(((...((((.(((((..-----(---------(.....)).......(((((((....))))))).....-...))))).)))).)))))))).............. ( -24.30) >DroPse_CAF1 187653 98 - 1 --CCCC-----UCGCCGUGUCAAUAAAUGA-----G---------CGCCAGGCAUUAAAUUGUAAUUUUUUAAUUACACACAA-AAGAUUUAAUGACUGCGGUGGUAAUAACACACAACA --....-----(((((((((((.(((((..-----(---------(.....)).......(((((((....))))))).....-...))))).)))).)))))))............... ( -21.80) >DroWil_CAF1 166341 109 - 1 -----------GCACCAUGUCAAUAAAUGAAGAGAGCGGACUAUUUGCCAAGCAUUAAAUUGUAAUUUUUUAAUUACACACAAAAAGAUUUAAUGACUGCGGUGGUAAUAACACACAACA -----------.((((..((((.(((((.....(.(((((...))))))...........(((((((....))))))).........))))).))))...))))................ ( -18.40) >DroYak_CAF1 165856 100 - 1 GACCGC-----ACGCCGUGUCAAUAAAUGA-----G---------CGCAAAGCAUUAAAUUGUAAUUUUUUAAUUACACACAA-AAGAUUUAAUGACUGCGGUGGUAAUAACACACAACA .(((((-----.(((...((((.(((((..-----(---------(.....)).......(((((((....))))))).....-...))))).)))).)))))))).............. ( -24.30) >DroAna_CAF1 178655 105 - 1 UACCGCCGUGUGCCCCUUGUCAAUAAAUGA-----G---------CGCAAAGCAUUAAAUUGUAAUUUUUUAAUUACGUACAA-AAUAUUUAAUGACUGUGGUGGUAAUAACACACAACA (((((((((((((.(.((.......)).).-----)---------)))).(((((((((((((((((....))))))).....-...)))))))).)).))))))))............. ( -24.10) >DroPer_CAF1 185049 98 - 1 --CCCC-----UCGCCGUGUCAAUAAAUGA-----G---------CGCCAGGCAUUAAAUUGUAAUUUUUUAAUUACACACAA-AAGAUUUAAUGACUGCGGUGGUAAUAACACACAACA --....-----(((((((((((.(((((..-----(---------(.....)).......(((((((....))))))).....-...))))).)))).)))))))............... ( -21.80) >consensus __CCGC_____ACGCCGUGUCAAUAAAUGA_____G_________CGCAAAGCAUUAAAUUGUAAUUUUUUAAUUACACACAA_AAGAUUUAAUGACUGCGGUGGUAAUAACACACAACA ............((((((((((.(((((..................((...)).......(((((((....))))))).........))))).)))).))))))................ (-17.42 = -17.67 + 0.25)

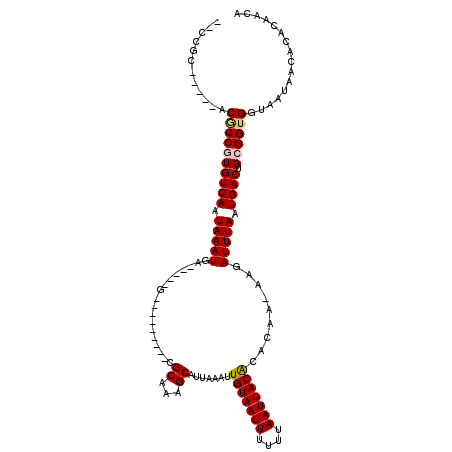
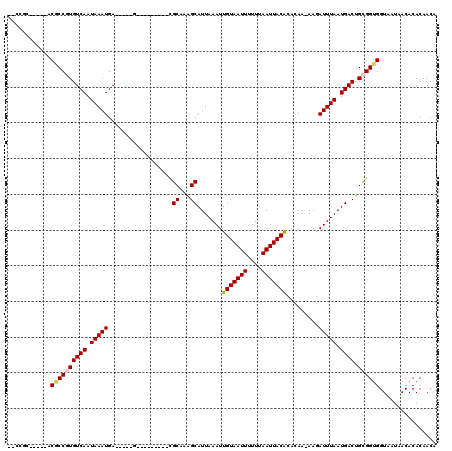
| Location | 4,730,960 – 4,731,052 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 87.56 |
| Mean single sequence MFE | -18.36 |
| Consensus MFE | -16.34 |
| Energy contribution | -17.14 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4730960 92 + 22407834 UGUGUAAUUAAAAAAUUACAAUUUAAUGCUUUGCGCUCAUUUAUUGACACGGCGUGCGGUCGUUGCAAAAGCAUUCA------CAUCCGCACCCAUAU .((((((((....)))).((((.....((.....))......))))))))((.((((((..(((((....)))...)------)..)))))))).... ( -22.50) >DroSec_CAF1 159810 82 + 1 UGUGUAAUUAAAAAAUUACAAUUUAAUGCCUUGCGCUCAUUUAUUGACACGGCGUGCGGUCGUUGCAAAAGCAUUCA------CAUCC---------- ..(((((((....)))))))....(((((.(((((.(((.....))))(((((.....))))).))))..)))))..------.....---------- ( -16.50) >DroSim_CAF1 155440 82 + 1 UGUGUAAUUAAAAAAUUACAAUUUAAUGCCUUGCGCUCAUUUAUUGACACGGCGUGCGGUCGUUGCAAAAGCAUUCA------CAUCC---------- ..(((((((....)))))))....(((((.(((((.(((.....))))(((((.....))))).))))..)))))..------.....---------- ( -16.50) >DroEre_CAF1 167424 88 + 1 UGUGUAAUUAAAAAAUUACAAUUUAAUGCUUUGCGCUCAUUUAUUGACACGGCGUGCGGUCGUUGCAAAAGCAUUCACAUUCACAUCC---------- ..(((((((....)))))))....(((((((((((.(((.....))))(((((.....))))).)).)))))))).............---------- ( -18.80) >DroYak_CAF1 165895 74 + 1 UGUGUAAUUAAAAAAUUACAAUUUAAUGCUUUGCGCUCAUUUAUUGACACGGCGUGCGGUCGUUGCUAAAGCAC------------------------ ..(((((((....)))))))......(((((((.(((((.....))).(((((.....))))).))))))))).------------------------ ( -17.50) >consensus UGUGUAAUUAAAAAAUUACAAUUUAAUGCUUUGCGCUCAUUUAUUGACACGGCGUGCGGUCGUUGCAAAAGCAUUCA______CAUCC__________ ..(((((((....)))))))....(((((((((((.(((.....))))(((((.....))))).)).))))))))....................... (-16.34 = -17.14 + 0.80)


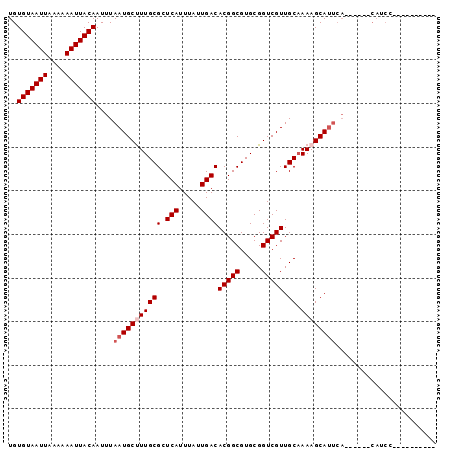
| Location | 4,730,960 – 4,731,052 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 87.56 |
| Mean single sequence MFE | -19.04 |
| Consensus MFE | -16.60 |
| Energy contribution | -16.40 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4730960 92 - 22407834 AUAUGGGUGCGGAUG------UGAAUGCUUUUGCAACGACCGCACGCCGUGUCAAUAAAUGAGCGCAAAGCAUUAAAUUGUAAUUUUUUAAUUACACA ((((((((((((.((------(...(((....)))))).)))))).))))))..........((.....)).......(((((((....))))))).. ( -27.00) >DroSec_CAF1 159810 82 - 1 ----------GGAUG------UGAAUGCUUUUGCAACGACCGCACGCCGUGUCAAUAAAUGAGCGCAAGGCAUUAAAUUGUAAUUUUUUAAUUACACA ----------.(((.------..(((((((((((.......)))((((((........))).))).))))))))..)))((((((....))))))... ( -17.30) >DroSim_CAF1 155440 82 - 1 ----------GGAUG------UGAAUGCUUUUGCAACGACCGCACGCCGUGUCAAUAAAUGAGCGCAAGGCAUUAAAUUGUAAUUUUUUAAUUACACA ----------.(((.------..(((((((((((.......)))((((((........))).))).))))))))..)))((((((....))))))... ( -17.30) >DroEre_CAF1 167424 88 - 1 ----------GGAUGUGAAUGUGAAUGCUUUUGCAACGACCGCACGCCGUGUCAAUAAAUGAGCGCAAAGCAUUAAAUUGUAAUUUUUUAAUUACACA ----------.(((((...((((..(((....)))..(((((.....)).)))..........))))..)))))....(((((((....))))))).. ( -17.70) >DroYak_CAF1 165895 74 - 1 ------------------------GUGCUUUAGCAACGACCGCACGCCGUGUCAAUAAAUGAGCGCAAAGCAUUAAAUUGUAAUUUUUUAAUUACACA ------------------------(((((((.((.......)).((((((........))).))).))))))).....(((((((....))))))).. ( -15.90) >consensus __________GGAUG______UGAAUGCUUUUGCAACGACCGCACGCCGUGUCAAUAAAUGAGCGCAAAGCAUUAAAUUGUAAUUUUUUAAUUACACA .......................((((((((.((.......)).((((((........))).))).))))))))....(((((((....))))))).. (-16.60 = -16.40 + -0.20)

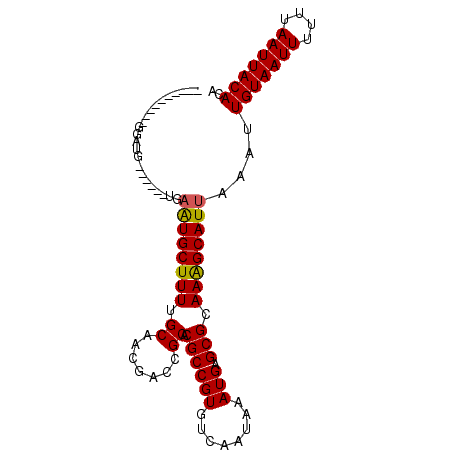
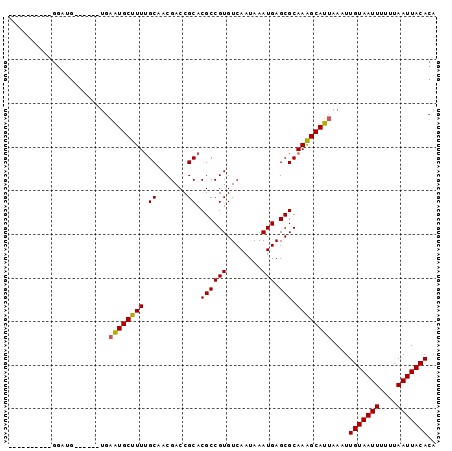
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:27 2006