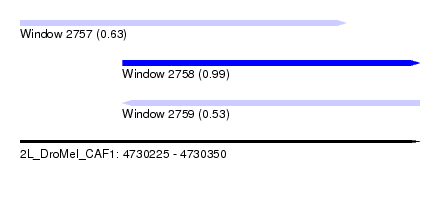
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,730,225 – 4,730,350 |
| Length | 125 |
| Max. P | 0.987011 |

| Location | 4,730,225 – 4,730,327 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -16.08 |
| Energy contribution | -15.92 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
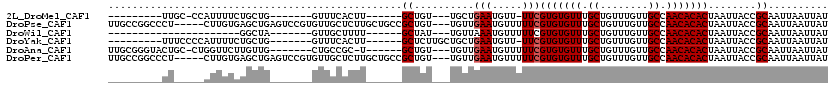
>2L_DroMel_CAF1 4730225 102 + 22407834 ---UUUUGC--UGGACGGACUUGGCUC-C--GCAGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAA-AACAUUCAGCA---ACAGC------AAG ---..((((--(((.((((......))-)--))..((.(((........)))........(((((((.((........)).)))))))...-........)).---.))))------)). ( -29.60) >DroPse_CAF1 186698 99 + 1 ----------------GUUCUCUGUUCCC--CAAGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAACAUUCAACA---ACAGCGGCAGCAAG ----------------((((((((((...--....((.(((........))).)).....(((((((.((........)).)))))))..............)---)))).)).)))... ( -21.30) >DroWil_CAF1 165363 92 + 1 ---UU---------------UUGGGUC-CUGGCAGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAACAUUUAACA---AUAGC------AAA ---..---------------.......-...((..((.(((........))).)).....(((((((.((........)).)))))))...............---...))------... ( -17.90) >DroYak_CAF1 165206 105 + 1 ---UUUUGG--CGGACGGACUUGGCUC-C--ACAGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAA-AACAUUCAGCAGCAAGAGC------AAG ---.((((.--....))))((((.(((-.--....((.(((........)))........(((((((.((........)).)))))))...-........)).....))))------))) ( -23.90) >DroAna_CAF1 177667 107 + 1 GGCUUUCACUAUAGAACGACUUCGUUC-C--GCAGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAACAUUCAACA---ACAGC------A-G (..((((......(((((....)))))-(--((((.............))))).......(((((((.((........)).)))))))))))..)........---.....------.-. ( -24.32) >DroPer_CAF1 184125 99 + 1 ----------------GUUCUCUGUUCCC--CAAGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAACAUUCAACA---ACAGCGGCAGCAAG ----------------((((((((((...--....((.(((........))).)).....(((((((.((........)).)))))))..............)---)))).)).)))... ( -21.30) >consensus ___UU___________GGACUUGGUUC_C__GCAGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAACAUUCAACA___ACAGC______AAG .......................(((.........((.(((........))).)).....(((((((.((........)).)))))))...........))).................. (-16.08 = -15.92 + -0.17)

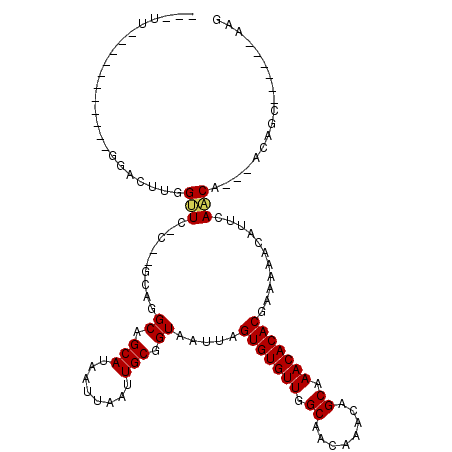
| Location | 4,730,257 – 4,730,350 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.42 |
| Mean single sequence MFE | -22.05 |
| Consensus MFE | -12.67 |
| Energy contribution | -12.53 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.987011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4730257 93 + 22407834 AUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAA-AACAUUCAGCA---ACAGC------AAGUGAAAC-------CAGCAGAAAAUGG-GCAA--------- ........(((((((.....(((((((.((........)).)))))))...-..((((..((.---...))------.))))..))-------).)))).......-....--------- ( -20.60) >DroPse_CAF1 186720 112 + 1 AUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAACAUUCAACA---ACAGCGGCAGCAAGAGCAACACGGACUCAGCUCACAAG-----AGGGCCGGCAA ........((((........(((((((.((........)).)))))))...............---....((((.....((((......).)))..(((....)-----)).)))))))) ( -26.70) >DroWil_CAF1 165384 82 + 1 AUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAACAUUUAACA---AUAGC------AAAAGCAAC-------UAGCC---------------------- ..........(((((.....(((((((.((........)).)))))))...............---...((------....)).))-------).)).---------------------- ( -15.00) >DroYak_CAF1 165238 97 + 1 AUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAA-AACAUUCAGCAGCAAGAGC------AAGUGAAAC-------CAGCAGAAAAUGGGGAAA--------- ........(((((((.....(((((((.((........)).)))))))...-....((((...((....))------...))))))-------).))))............--------- ( -21.10) >DroAna_CAF1 177704 102 + 1 AUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAACAUUCAACA---ACAGC------A-GCGGCAG-------CAACAAGAACCAG-GCAGUACCCGCAA ........((((((..(((.(((((((.((........)).)))))))...............---...((------.-..((...-------(.....)..))..-)))))..)))))) ( -22.20) >DroPer_CAF1 184147 112 + 1 AUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAACAUUCAACA---ACAGCGGCAGCAAGAGCAACACGGACUCAGCUCACAAG-----AGGGCCGGCAA ........((((........(((((((.((........)).)))))))...............---....((((.....((((......).)))..(((....)-----)).)))))))) ( -26.70) >consensus AUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAACAUUCAACA___ACAGC______AAGAGCAAC_______CAGCACAAAAG_____AG_________ ..........((........(((((((.((........)).)))))))(((.....)))..........))................................................. (-12.67 = -12.53 + -0.14)

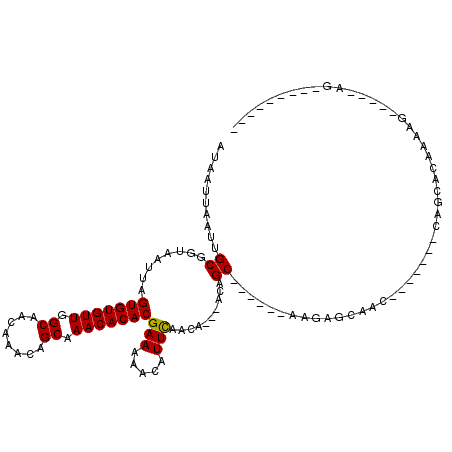
| Location | 4,730,257 – 4,730,350 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.42 |
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -13.20 |
| Energy contribution | -13.37 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
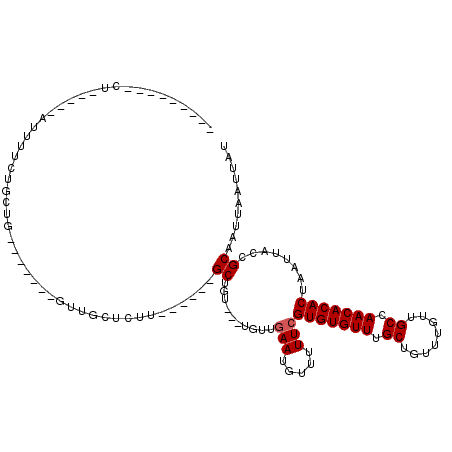
>2L_DroMel_CAF1 4730257 93 - 22407834 ---------UUGC-CCAUUUUCUGCUG-------GUUUCACUU------GCUGU---UGCUGAAUGUU-UUCGUGUGUUUGCUGUUUGUUGCCAACACACUAAUUACCGCAAUUAAUUAU ---------((((-.(((((...((.(-------((.......------)))..---.)).)))))..-...(((((((.((........)).)))))))........))))........ ( -19.40) >DroPse_CAF1 186720 112 - 1 UUGCCGGCCCU-----CUUGUGAGCUGAGUCCGUGUUGCUCUUGCUGCCGCUGU---UGUUGAAUGUUUUUCGUGUGUUUGCUGUUUGUUGCCAACACACUAAUUACCGCAAUUAAUUAU ((((((((.(.-----.....)(((.((((.......))))..)))))))....---........((..((.(((((((.((........)).))))))).))..)).))))........ ( -25.40) >DroWil_CAF1 165384 82 - 1 ----------------------GGCUA-------GUUGCUUUU------GCUAU---UGUUAAAUGUUUUUCGUGUGUUUGCUGUUUGUUGCCAACACACUAAUUACCGCAAUUAAUUAU ----------------------...((-------((((((((.------((...---.)).))).((..((.(((((((.((........)).))))))).))..)).)))))))..... ( -18.60) >DroYak_CAF1 165238 97 - 1 ---------UUUCCCCAUUUUCUGCUG-------GUUUCACUU------GCUCUUGCUGCUGAAUGUU-UUCGUGUGUUUGCUGUUUGUUGCCAACACACUAAUUACCGCAAUUAAUUAU ---------.............(((.(-------((((((...------((....))...))))....-...(((((((.((........)).))))))).....))))))......... ( -20.60) >DroAna_CAF1 177704 102 - 1 UUGCGGGUACUGC-CUGGUUCUUGUUG-------CUGCCGC-U------GCUGU---UGUUGAAUGUUUUUCGUGUGUUUGCUGUUUGUUGCCAACACACUAAUUACCGCAAUUAAUUAU ((((((.......-...((((.....(-------(.((...-.------)).))---....)))).......(((((((.((........)).)))))))......))))))........ ( -25.10) >DroPer_CAF1 184147 112 - 1 UUGCCGGCCCU-----CUUGUGAGCUGAGUCCGUGUUGCUCUUGCUGCCGCUGU---UGUUGAAUGUUUUUCGUGUGUUUGCUGUUUGUUGCCAACACACUAAUUACCGCAAUUAAUUAU ((((((((.(.-----.....)(((.((((.......))))..)))))))....---........((..((.(((((((.((........)).))))))).))..)).))))........ ( -25.40) >consensus _________CU_____AUUUUCUGCUG_______GUUGCUCUU______GCUGU___UGUUGAAUGUUUUUCGUGUGUUUGCUGUUUGUUGCCAACACACUAAUUACCGCAAUUAAUUAU .................................................((..........(((.....)))(((((((.((........)).)))))))........)).......... (-13.20 = -13.37 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:23 2006