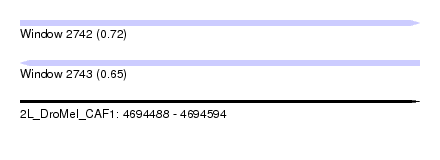
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,694,488 – 4,694,594 |
| Length | 106 |
| Max. P | 0.722408 |

| Location | 4,694,488 – 4,694,594 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.00 |
| Mean single sequence MFE | -15.88 |
| Consensus MFE | -10.80 |
| Energy contribution | -10.68 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4694488 106 + 22407834 CAAGGAAUACAUAUUUAUUUAUGCACAGAAUUUUUCUCUGUAUUCCCAAAAGGGGAAAUAAACAUCAGACCAACACAGAAUUCCACUCAAAACAAUUAUGAAUGAA ...(((((.((((......)))).(((((.......))))).(((((.....)))))......................)))))...................... ( -16.40) >DroSec_CAF1 124013 106 + 1 GAAGAAAUACAUAUUUAUUUAUGCACACAAUUUUUCUCUGUAUUCCCGAAAGGGGAAAUAAGCAUCAGACCAACACAGAAUUCCACUCAAAACAAUUAUGAAUGAA ((((((...((((......)))).......))))))(((((.(((((....)))))...........(.....))))))....((.(((.........))).)).. ( -14.60) >DroSim_CAF1 116902 106 + 1 AAAGAAAUACAUAUUUAUUUAUGCACACAAUUUUUCUCUGUAUUCCCGAAAGGGGAAAUAAACAUCAGACCAACACAGAAUUCCACUCAAAACAAUUAUGAAUGAA .............((((((((((.............(((((.(((((....)))))......(....)......))))).................)))))))))) ( -14.11) >DroEre_CAF1 127862 85 + 1 ---------------------UGCACAGCAUUUCGCUGUGUAUUCCCGAAAGUGGAAAUAAACAUCAGGCCAACACACAAUUUCACUCAAAACAAUUAUGAAUGAA ---------------------((((((((.....))))))))(((((....).))))........................((((.(((.........))).)))) ( -19.50) >DroYak_CAF1 128617 85 + 1 ---------------------CGUACAGAAUUUUUCUCUGUAUUCCCGAAAGGGGAAAUAAACAUCAGGCCAACACAGAAUUCCACUCAAAACAAUUAUGAAUGAA ---------------------..((((((.......))))))(((((....)))))...........................((.(((.........))).)).. ( -14.80) >consensus _AAG_AAUACAUAUUUAUUUAUGCACAGAAUUUUUCUCUGUAUUCCCGAAAGGGGAAAUAAACAUCAGACCAACACAGAAUUCCACUCAAAACAAUUAUGAAUGAA ....................................(((((.(((((....)))))...........(.....))))))....((.(((.........))).)).. (-10.80 = -10.68 + -0.12)

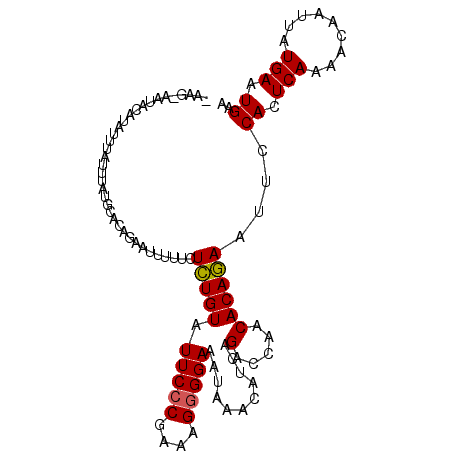

| Location | 4,694,488 – 4,694,594 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.00 |
| Mean single sequence MFE | -21.77 |
| Consensus MFE | -18.70 |
| Energy contribution | -17.82 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4694488 106 - 22407834 UUCAUUCAUAAUUGUUUUGAGUGGAAUUCUGUGUUGGUCUGAUGUUUAUUUCCCCUUUUGGGAAUACAGAGAAAAAUUCUGUGCAUAAAUAAAUAUGUAUUCCUUG ((((((((.........)))))))).(((((((((.....((........))(((....)))))))))))).........(((((((......)))))))...... ( -22.90) >DroSec_CAF1 124013 106 - 1 UUCAUUCAUAAUUGUUUUGAGUGGAAUUCUGUGUUGGUCUGAUGCUUAUUUCCCCUUUCGGGAAUACAGAGAAAAAUUGUGUGCAUAAAUAAAUAUGUAUUUCUUC ((((((((.........)))))))).((((((((((((.....)))......(((....)))))))))))).........(((((((......)))))))...... ( -22.40) >DroSim_CAF1 116902 106 - 1 UUCAUUCAUAAUUGUUUUGAGUGGAAUUCUGUGUUGGUCUGAUGUUUAUUUCCCCUUUCGGGAAUACAGAGAAAAAUUGUGUGCAUAAAUAAAUAUGUAUUUCUUU ((((((((.........)))))))).(((((((((.....((........))(((....)))))))))))).........(((((((......)))))))...... ( -22.30) >DroEre_CAF1 127862 85 - 1 UUCAUUCAUAAUUGUUUUGAGUGAAAUUGUGUGUUGGCCUGAUGUUUAUUUCCACUUUCGGGAAUACACAGCGAAAUGCUGUGCA--------------------- ((((((((.........))))))))..(((((((...(((((.((........))..))))).)))))))(((........))).--------------------- ( -21.40) >DroYak_CAF1 128617 85 - 1 UUCAUUCAUAAUUGUUUUGAGUGGAAUUCUGUGUUGGCCUGAUGUUUAUUUCCCCUUUCGGGAAUACAGAGAAAAAUUCUGUACG--------------------- ((((((((.........)))))))).((((((((...(((((...............))))).))))))))..............--------------------- ( -19.86) >consensus UUCAUUCAUAAUUGUUUUGAGUGGAAUUCUGUGUUGGUCUGAUGUUUAUUUCCCCUUUCGGGAAUACAGAGAAAAAUUCUGUGCAUAAAUAAAUAUGUAUU_CUU_ ((((((((.........)))))))).(((((((((..(((((...............))))))))))))))................................... (-18.70 = -17.82 + -0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:08 2006