
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,691,349 – 4,691,484 |
| Length | 135 |
| Max. P | 0.959067 |

| Location | 4,691,349 – 4,691,453 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -26.16 |
| Consensus MFE | -15.57 |
| Energy contribution | -16.22 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
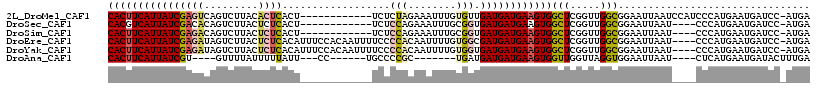
>2L_DroMel_CAF1 4691349 104 - 22407834 CACUUCAUUAUCGAGUCAGUCUUACACUCACU------------UCUCUAGAAAUUUGUGUUGAUGAUGAAGUGGCUCGGUUGGCGGAAUUAAUCCAUCCCAUGAAUGAUCC-AUGA (((((((((((((((.....)))((((....(------------((....)))....)))).))))))))))))(((.....)))(((.....)))....((((.......)-))). ( -26.60) >DroSec_CAF1 120942 100 - 1 CACGUCAUUAUCGACACAGUCUUACUCUCACU------------UCUCCAGAAAUUUGCGGUGAUGAUGAAGUGGCUCGGUUGGCGGAAUUAAU----CCCAUGAAUGAUCC-AUGA ..(((((((((((......(((..........------------.....)))......)))))))))))..((((.(((....(.(((.....)----))).....))).))-)).. ( -22.56) >DroSim_CAF1 113821 100 - 1 CACUUCAUUAUCGAGACAGUCUUACUCUCACU------------UCUCCAGAAAUUUGCGGUGAUGAUGAAGUGGCUCGGUUGGCGGAAUUAAU----CCCAUGAAUGAUCC-AUGA ((((((((((((((((.........))))..(------------((....))).........))))))))))))(((.....)))(((.....)----))((((.......)-))). ( -25.00) >DroEre_CAF1 124809 112 - 1 CACUUCAUUAUCGAGAUAGUCUUACUCUCACAUUUCCACAAUUUUCCCCACAAUUUUGUGGCGAUGAUGAAGUGGCUCGGUUGGCGGAAUUAAU----CCCAUGAAUGAUCC-AUGA ((((((((((((((((.........))))......((((((..((......))..)))))).))))))))))))(((.....)))(((.....)----))((((.......)-))). ( -31.70) >DroYak_CAF1 125587 112 - 1 CACUUCAUUAUCGAGAUAGUCUUACUCUCACAUUUCCACAAUUUUCCCCACAAUUUUGUGGUGAUGAUGAAGUGGCUCGGUUGGCGGAAUUAAU----CCCAUGAAUGAUCC-AUGA ((((((((((((((((.........))))......((((((..((......))..)))))).))))))))))))(((.....)))(((.....)----))((((.......)-))). ( -31.60) >DroAna_CAF1 138062 93 - 1 CACUUCAUUAUCGU----GUUUUAUUUUUAUU---CC------UGCCCCGC-------UGAUGAUGAUGAAGUGGUUGGUUAGGUGGAAUUAAU----CUCAUGAAUGAUACUUUGA ....((((..((((----(....(((...(((---((------.(((..((-------..((.((......)).))..))..)))))))).)))----..)))))))))........ ( -19.50) >consensus CACUUCAUUAUCGAGACAGUCUUACUCUCACU____________UCCCCACAAAUUUGUGGUGAUGAUGAAGUGGCUCGGUUGGCGGAAUUAAU____CCCAUGAAUGAUCC_AUGA ((((((((((((((((.........))))..................(((........))).))))))))))))(((.....)))................................ (-15.57 = -16.22 + 0.64)
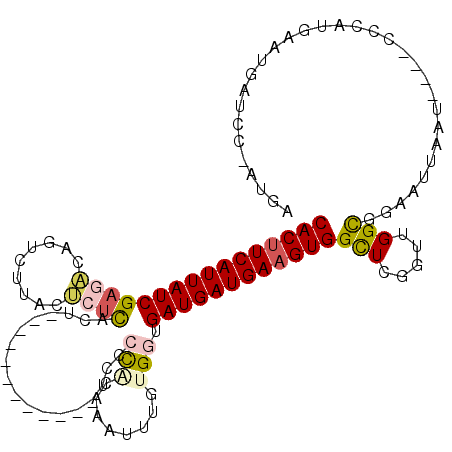

| Location | 4,691,387 – 4,691,484 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.08 |
| Mean single sequence MFE | -21.28 |
| Consensus MFE | -15.23 |
| Energy contribution | -16.15 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4691387 97 - 22407834 AGAAACCCCUAAAUCUU-----UGUCUAUUUCCGUACACUUCAUUAUCGAGUCAGUCUUACACUCACU------------UCUCUAGAAAUUUGUGUUGAUGAUGAAGUGGCUC .................-----..............(((((((((((((((.....)))((((....(------------((....)))....)))).)))))))))))).... ( -20.80) >DroSec_CAF1 120976 97 - 1 AGAAACCCCUAAAUCUU-----UGUCUAUUUCCGUACACGUCAUUAUCGACACAGUCUUACUCUCACU------------UCUCCAGAAAUUUGCGGUGAUGAUGAAGUGGCUC .......((.....(((-----..((.(((.(((((...(((......)))................(------------((....)))...))))).)))))..))).))... ( -16.20) >DroSim_CAF1 113855 97 - 1 AGAAACCCCUAAAUCUU-----UGUCUAUUUCCGUACACUUCAUUAUCGAGACAGUCUUACUCUCACU------------UCUCCAGAAAUUUGCGGUGAUGAUGAAGUGGCUC .................-----..............((((((((((((((((.........))))..(------------((....))).........)))))))))))).... ( -20.10) >DroEre_CAF1 124843 109 - 1 AGAAACCCCUAAAUCUU-----UGUCUAUUUCCGUACACUUCAUUAUCGAGAUAGUCUUACUCUCACAUUUCCACAAUUUUCCCCACAAUUUUGUGGCGAUGAUGAAGUGGCUC .................-----..............((((((((((((((((.........))))......((((((..((......))..)))))).)))))))))))).... ( -26.80) >DroYak_CAF1 125621 109 - 1 AGAAAUCCCUAAAUCUU-----UGUCUAUUUCCGUACACUUCAUUAUCGAGAUAGUCUUACUCUCACAUUUCCACAAUUUUCCCCACAAUUUUGUGGUGAUGAUGAAGUGGCUC .................-----..............((((((((((((((((.........))))......((((((..((......))..)))))).)))))))))))).... ( -26.70) >DroAna_CAF1 138097 94 - 1 AGAAAUGCUUAAAUCUUCUGCCUGUCCAUUUUCCUUCACUUCAUUAUCGU----GUUUUAUUUUUAUU---CC------UGCCCCGC-------UGAUGAUGAUGAAGUGGUUG .((...((...........))...)).........(((((((((((((((----..............---..------........-------..)))))))))))))))... ( -17.07) >consensus AGAAACCCCUAAAUCUU_____UGUCUAUUUCCGUACACUUCAUUAUCGAGACAGUCUUACUCUCACU____________UCCCCACAAAUUUGUGGUGAUGAUGAAGUGGCUC ....................................((((((((((((((((.........))))..................(((........))).)))))))))))).... (-15.23 = -16.15 + 0.92)

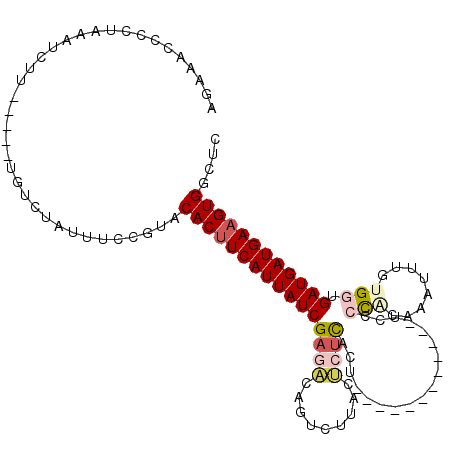
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:06 2006