
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,683,818 – 4,683,932 |
| Length | 114 |
| Max. P | 0.796090 |

| Location | 4,683,818 – 4,683,932 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 71.73 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -17.91 |
| Energy contribution | -18.42 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
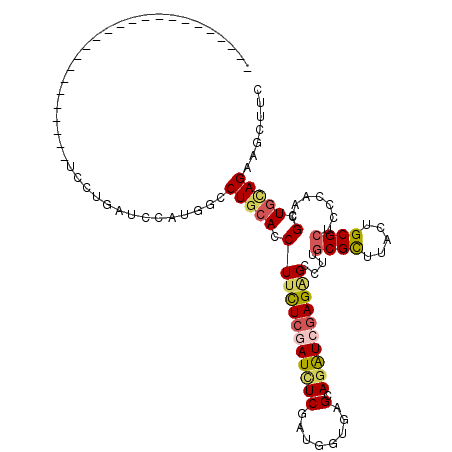
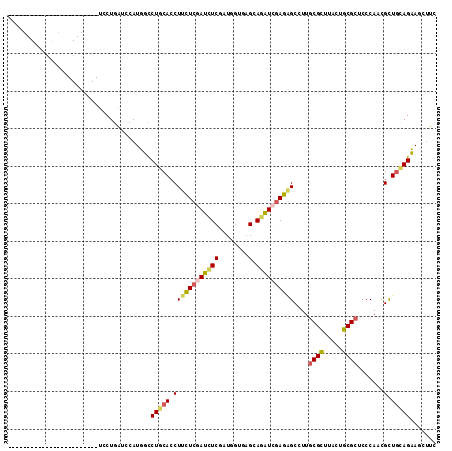
>2L_DroMel_CAF1 4683818 114 + 22407834 UCAUGAGACUGUUUGUGCUCUGGCUCCUGAUCCAUUUCCUGGACCUUUUCAAUUUCGAUGGUGAGGAGAUCGAGAGCCUUGCGCUUACUGCGCUCCCAUUGCUGGAGAAGCUUC ....(((...((..((((...(((((.(((((....((((.(.((...((......)).))).))))))))).)))))..))))..))....))).....(((.....)))... ( -35.20) >DroVir_CAF1 130526 90 + 1 ------------------------UCCUGAUCCAUGGUCUGCACCUUCUCGAUCUCGAUGGUGAGCAAAUCCAGGGCCUUGCGUUUGGUGCGCUCCCAACGCUGCAGCAUCUUU ------------------------...........((((((((.(...(((....)))(((.((((....(((((((...)).)))))...)))))))..).))))).)))... ( -23.60) >DroGri_CAF1 116221 90 + 1 ------------------------UCCUGAUCCAUGGCCUGCACCUUCUCGAUCUCAAUGGUAAGCAGAUCGAGAGCCUUCCGCUUGGAGCGUUCCCAACGUUGCAGCAUCUUC ------------------------...........(((........((((((((((........).))))))))))))....(((.(.(((((.....))))).))))...... ( -23.70) >DroEre_CAF1 117177 108 + 1 UCAUGAG------CCUGCUCUUGCUCCUGAUCCAUUUCCUGCACCUUCUCGAUCUCAGUUGUUAGGAGAUCGAGAGCCUUGCGCUUACUGCGCUCCCAUCGCUGUAGAAGCCUC (((.(((------(........)))).)))..........(((..(((((((((((.........)))))))))))...)))((((.((((((.......)).))))))))... ( -34.60) >DroYak_CAF1 117825 114 + 1 UCAUGAGUCGGCUCCUGCUCUUGCUCCUGAUCCAUUUCCUGCACCUUCUCAAUCUCGAUGGUCAGGAGGUCGAGAGCCUUGCGCUUACUGCGCUCCCAUUGCUGCAGAAGCUUC .........((((.((((....((((.(((((....(((((.(((.((........)).))))))))))))).))))...((((.....))))..........)))).)))).. ( -38.70) >DroMoj_CAF1 138238 90 + 1 ------------------------UCCUGGUCCAUGGUCUGCAGCUUCUCGAUCUCGAGCAUGAGCAGGUCGAGCGCCUUGCGCUUGCUGCGCUCCCAACGCUGCAGCAUCUUG ------------------------........((.((((((((((..((((....))))...((((.((.(((((((...)))))))))..)))).....))))))).))).)) ( -36.10) >consensus ________________________UCCUGAUCCAUGGCCUGCACCUUCUCGAUCUCGAUGGUGAGCAGAUCGAGAGCCUUGCGCUUACUGCGCUCCCAACGCUGCAGAAGCUUC ......................................(((((.((((((((((((........).))))))))))....((((.....)))).......).)))))....... (-17.91 = -18.42 + 0.50)

| Location | 4,683,818 – 4,683,932 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 71.73 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -16.68 |
| Energy contribution | -18.02 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
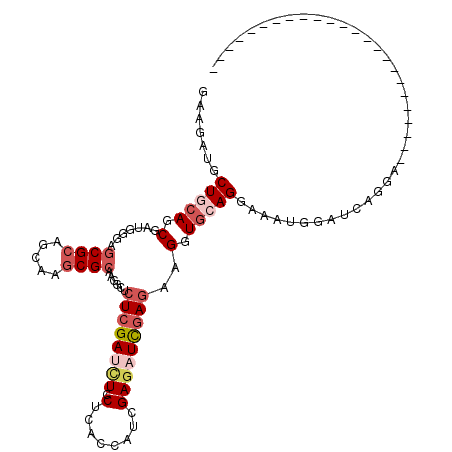
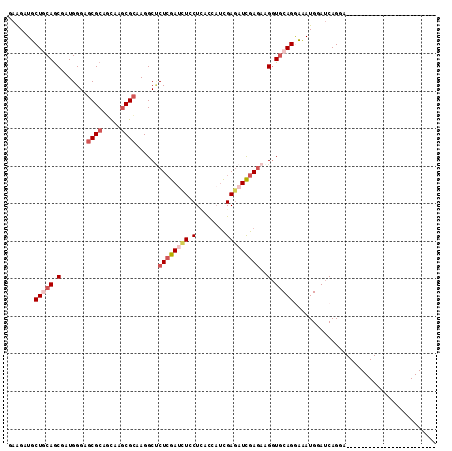
>2L_DroMel_CAF1 4683818 114 - 22407834 GAAGCUUCUCCAGCAAUGGGAGCGCAGUAAGCGCAAGGCUCUCGAUCUCCUCACCAUCGAAAUUGAAAAGGUCCAGGAAAUGGAUCAGGAGCCAGAGCACAAACAGUCUCAUGA ...((((.((((....)))).((((.....))))..(((((..((((((((.(((.(((....)))...)))..))))....))))..))))).))))................ ( -35.30) >DroVir_CAF1 130526 90 - 1 AAAGAUGCUGCAGCGUUGGGAGCGCACCAAACGCAAGGCCCUGGAUUUGCUCACCAUCGAGAUCGAGAAGGUGCAGACCAUGGAUCAGGA------------------------ ...((((((...(((((((.......)).)))))..)))(((((.(((((..(((.(((....)))...))))))))))).)))))....------------------------ ( -28.20) >DroGri_CAF1 116221 90 - 1 GAAGAUGCUGCAACGUUGGGAACGCUCCAAGCGGAAGGCUCUCGAUCUGCUUACCAUUGAGAUCGAGAAGGUGCAGGCCAUGGAUCAGGA------------------------ .......(((((.((((.(((....))).))))......(((((((((.(........))))))))))...))))).((........)).------------------------ ( -27.10) >DroEre_CAF1 117177 108 - 1 GAGGCUUCUACAGCGAUGGGAGCGCAGUAAGCGCAAGGCUCUCGAUCUCCUAACAACUGAGAUCGAGAAGGUGCAGGAAAUGGAUCAGGAGCAAGAGCAGG------CUCAUGA ...((((((............((((.....)))).....((((((((((.........)))))))))).(((.((.....)).)))))))))..(((....------))).... ( -35.70) >DroYak_CAF1 117825 114 - 1 GAAGCUUCUGCAGCAAUGGGAGCGCAGUAAGCGCAAGGCUCUCGACCUCCUGACCAUCGAGAUUGAGAAGGUGCAGGAAAUGGAUCAGGAGCAAGAGCAGGAGCCGACUCAUGA ...((((((((..........((((.....))))...((((..((((((((((((.((........)).))).)))))...)).))..))))....)))))))).......... ( -44.20) >DroMoj_CAF1 138238 90 - 1 CAAGAUGCUGCAGCGUUGGGAGCGCAGCAAGCGCAAGGCGCUCGACCUGCUCAUGCUCGAGAUCGAGAAGCUGCAGACCAUGGACCAGGA------------------------ .......(((((((((.((((((((.((....))...))))))..)).)).....((((....))))..))))))).((........)).------------------------ ( -34.10) >consensus GAAGAUGCUGCAGCGAUGGGAGCGCAGCAAGCGCAAGGCUCUCGAUCUCCUCACCAUCGAGAUCGAGAAGGUGCAGGAAAUGGAUCAGGA________________________ .......(((((.(.......((((.....))))......((((((((.(........)))))))))..).)))))...................................... (-16.68 = -18.02 + 1.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:04 2006