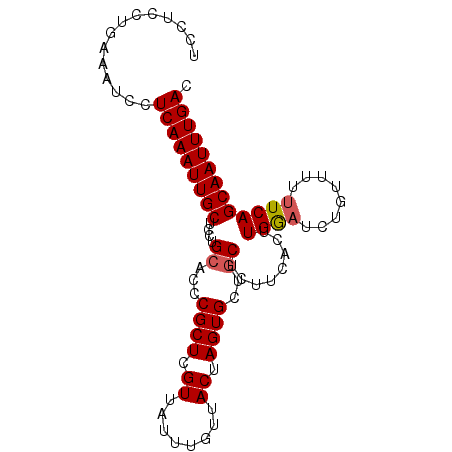
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,675,940 – 4,676,032 |
| Length | 92 |
| Max. P | 0.950064 |

| Location | 4,675,940 – 4,676,032 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 95.65 |
| Mean single sequence MFE | -17.20 |
| Consensus MFE | -14.72 |
| Energy contribution | -14.96 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4675940 92 + 22407834 UCCUCCAGAAAUCCUCAAAUUGCUCCUGCACCCGCUCGUUAUUUGUUACUAGUGCUGCUCUUCACUUGAAUCUGUUUUUUCAGCAAUUUGAC ..............(((((((((....(((..((((.((........)).)))).)))........((((........))))))))))))). ( -17.80) >DroSec_CAF1 104645 92 + 1 UCCUCCUGAAAUCCUCAAAUUGCUCCUGCACCCGCUCGUUAUUUGUUACUAGUGCUGCUCUUCACUUGGAUCUGUUUUUUCAGCAAUUUGAC ..............(((((((((....(((..((((.((........)).)))).)))........((((........))))))))))))). ( -17.30) >DroSim_CAF1 96430 92 + 1 UCCUGCUGAAAUCCUCAAAUUGCUCCUGCACCCGCUCGUUAUUUGUUACUAGUGCUGCUCUUCACUUGGAUCUGUUUUUUCAGCAAUUUGAC ..............(((((((((....(((..((((.((........)).)))).)))........((((........))))))))))))). ( -17.30) >DroEre_CAF1 108351 91 + 1 UCCUCCUGAAAUCCUCAAAUUGCU-CUGCACCCGCUCGUUAUUUGUUACUAGUGCCGCUCUUCACUUGGAUCUGUUUUUUCAGCAAUUUGAC ..............(((((((((.-..((...((((.((........)).))))..))........((((........))))))))))))). ( -16.20) >DroYak_CAF1 108669 92 + 1 UCCUGCUGAAAUCCUCAAAUUGCUCCUGCACCCGCUCGUUAUUUGUUACUAGUGCCACUACUCACUUGGUUCUGUUUUUUCAGCAAUUUGAC ...((((((((....(((((.((....((....))..)).)))))....(((.((((.........)))).)))...))))))))....... ( -17.40) >consensus UCCUCCUGAAAUCCUCAAAUUGCUCCUGCACCCGCUCGUUAUUUGUUACUAGUGCUGCUCUUCACUUGGAUCUGUUUUUUCAGCAAUUUGAC ..............(((((((((....((...((((.((........)).))))..))........((((........))))))))))))). (-14.72 = -14.96 + 0.24)


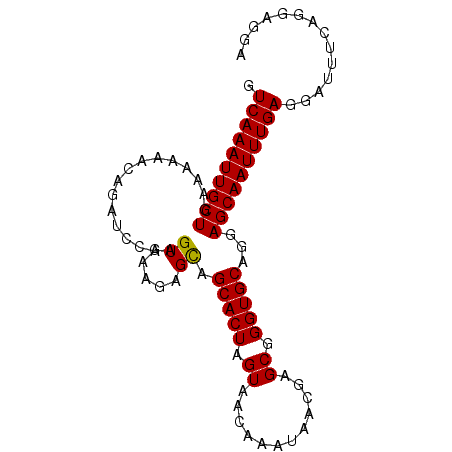
| Location | 4,675,940 – 4,676,032 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 95.65 |
| Mean single sequence MFE | -19.40 |
| Consensus MFE | -17.86 |
| Energy contribution | -17.70 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4675940 92 - 22407834 GUCAAAUUGCUGAAAAAACAGAUUCAAGUGAAGAGCAGCACUAGUAACAAAUAACGAGCGGGUGCAGGAGCAAUUUGAGGAUUUCUGGAGGA .((((((((((.........(.(((....)))...).(((((.((............)).)))))...)))))))))).............. ( -19.50) >DroSec_CAF1 104645 92 - 1 GUCAAAUUGCUGAAAAAACAGAUCCAAGUGAAGAGCAGCACUAGUAACAAAUAACGAGCGGGUGCAGGAGCAAUUUGAGGAUUUCAGGAGGA .(((((((((((......))..(((..((.....)).(((((.((............)).))))).)))))))))))).............. ( -19.40) >DroSim_CAF1 96430 92 - 1 GUCAAAUUGCUGAAAAAACAGAUCCAAGUGAAGAGCAGCACUAGUAACAAAUAACGAGCGGGUGCAGGAGCAAUUUGAGGAUUUCAGCAGGA ......(((((((((...(((((((..((.....)).(((((.((............)).))))).)))....))))....))))))))).. ( -20.60) >DroEre_CAF1 108351 91 - 1 GUCAAAUUGCUGAAAAAACAGAUCCAAGUGAAGAGCGGCACUAGUAACAAAUAACGAGCGGGUGCAG-AGCAAUUUGAGGAUUUCAGGAGGA .((((((((((................((.....)).(((((.((............)).)))))..-)))))))))).............. ( -18.70) >DroYak_CAF1 108669 92 - 1 GUCAAAUUGCUGAAAAAACAGAACCAAGUGAGUAGUGGCACUAGUAACAAAUAACGAGCGGGUGCAGGAGCAAUUUGAGGAUUUCAGCAGGA .((((((((((.......((..(((....).))..))(((((.((............)).)))))...)))))))))).............. ( -18.80) >consensus GUCAAAUUGCUGAAAAAACAGAUCCAAGUGAAGAGCAGCACUAGUAACAAAUAACGAGCGGGUGCAGGAGCAAUUUGAGGAUUUCAGGAGGA .((((((((((................((.....)).(((((.((............)).)))))...)))))))))).............. (-17.86 = -17.70 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:01 2006