| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
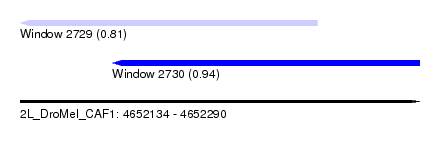
| Location | 4,652,134 – 4,652,290 |
| Length | 156 |
| Max. P | 0.939512 |

| Location | 4,652,134 – 4,652,250 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -23.25 |
| Consensus MFE | -17.58 |
| Energy contribution | -18.32 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4652134 116 - 22407834 GCUCGUAAAUAUUUUAUGUACGCCCUGAAAACUUCUAUUCAGGCCUACAUAAAAGGAAAUACAGUUUAGCACAUAAAUCAAGACUUGUACUCACGGUCUUGCCAUCAAAGUUAUGA ..((((((...(((((((((.(((.((((........))))))).)))))))))((.......(((((.....)))))(((((((((....)).))))))))).......)))))) ( -27.10) >DroSec_CAF1 88439 116 - 1 GCUCGUAAAUAUUUUAUGUACGCCCUGAAAACUUCUAUUCAGGCCUACAUAAAAGAAAAUACAGUUUGGCACAUAAAUCAAGGCUUGUACUCACGGUCUUGCCAUCAAAGUUAUGA ..((((((...(((((((((.(((.((((........))))))).)))))))))............(((((.........(((((((....)).))))))))))......)))))) ( -26.60) >DroSim_CAF1 80025 116 - 1 GCUCGUAAAUAUUUUAUGUACGCCCUGAAAACUUCUAUUCAGGCCUACAUAAAAGAAAAUACAGUUUUUCACAUAAAUCAAGGCUUGUACUCACGGUCUUGCCAUCAAAGUUAUGA ..((((((...(((((((((.(((.((((........))))))).)))))))))(((((......)))))........(((((((((....)).))))))).........)))))) ( -27.00) >DroEre_CAF1 91816 111 - 1 ACUCGUAAACAUUUUAUAUAUGCCCUGAAAACUUCUAUUCACGCCAACAUAA-----AAUACAGCAUAGCAAAUAAAUCAAGAAUAGUACUCACAGUUCUGCCAUCAAAGUUGUGG ..........(((((((....((..((((........)))).))....))))-----)))....((((((..........(((((.((....)).))))).........)))))). ( -12.31) >consensus GCUCGUAAAUAUUUUAUGUACGCCCUGAAAACUUCUAUUCAGGCCUACAUAAAAGAAAAUACAGUUUAGCACAUAAAUCAAGACUUGUACUCACGGUCUUGCCAUCAAAGUUAUGA ..((((((...(((((((((.(((.((((........))))))).)))))))))........................(((((((.((....))))))))).........)))))) (-17.58 = -18.32 + 0.75)


| Location | 4,652,170 – 4,652,290 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -22.73 |
| Consensus MFE | -18.42 |
| Energy contribution | -20.68 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4652170 120 - 22407834 CUCGACUAAGAACAUUCAGAUUUAGAUGUUUUCCUUAUAGGCUCGUAAAUAUUUUAUGUACGCCCUGAAAACUUCUAUUCAGGCCUACAUAAAAGGAAAUACAGUUUAGCACAUAAAUCA ..................((((((..(((........((((((.(((....(((((((((.(((.((((........))))))).))))))))).....))))))))))))..)))))). ( -26.40) >DroSec_CAF1 88475 120 - 1 CUCAGCUAAGAACAUUCAGAUUUAGAUGUUUUCCUUAUAGGCUCGUAAAUAUUUUAUGUACGCCCUGAAAACUUCUAUUCAGGCCUACAUAAAAGAAAAUACAGUUUGGCACAUAAAUCA ....((((((((((((........)))))))).......((((.(((....(((((((((.(((.((((........))))))).))))))))).....))))))))))).......... ( -26.60) >DroSim_CAF1 80061 120 - 1 CUCAGCUAAGAACUUUUAGAUUUAGAUGUUUUCCUUAUUGGCUCGUAAAUAUUUUAUGUACGCCCUGAAAACUUCUAUUCAGGCCUACAUAAAAGAAAAUACAGUUUUUCACAUAAAUCA ..................((((((.(((....((.....))..))).....(((((((((.(((.((((........))))))).)))))))))(((((......)))))...)))))). ( -22.90) >DroEre_CAF1 91852 112 - 1 CUUAGCUA---ACUAUCAGAUUUAGAUGUUUUCUUUAUGGACUCGUAAACAUUUUAUAUAUGCCCUGAAAACUUCUAUUCACGCCAACAUAA-----AAUACAGCAUAGCAAAUAAAUCA ....((((---.......((..((((.((((((((((((....))))))(((.......)))....)))))).)))).))..((........-----......)).)))).......... ( -15.04) >consensus CUCAGCUAAGAACAUUCAGAUUUAGAUGUUUUCCUUAUAGGCUCGUAAAUAUUUUAUGUACGCCCUGAAAACUUCUAUUCAGGCCUACAUAAAAGAAAAUACAGUUUAGCACAUAAAUCA ..................((((((..(((........((((((.(((....(((((((((.(((.((((........))))))).))))))))).....))))))))))))..)))))). (-18.42 = -20.68 + 2.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:56 2006