| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,650,411 – 4,650,508 |
| Length | 97 |
| Max. P | 0.905383 |

| Location | 4,650,411 – 4,650,508 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.09 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -20.38 |
| Energy contribution | -20.02 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
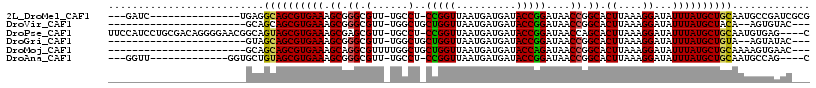
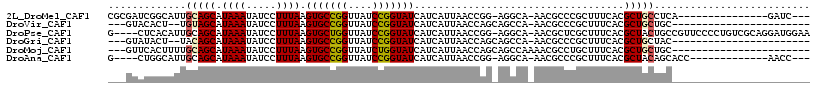
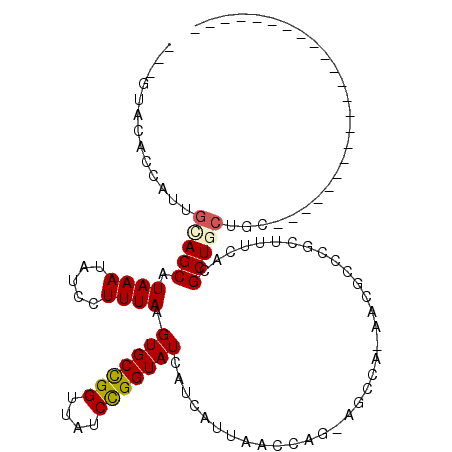
>2L_DroMel_CAF1 4650411 97 + 22407834 ---GAUC---------------UGAGGCAGCGUGAAAGCGGGCGUU-UGCCU-CCGGUUAAUGAUGAUACCGGAUAACCGGCACUUAAAGGAUAUUUAUGCUGCAAUGCCGAUCGCG ---((((---------------.(..((((((((((...((((...-.))))-(((((((.((.......))..))))))).............))))))))))....).))))... ( -32.00) >DroVir_CAF1 90903 88 + 1 -----------------------GCAGCAGCGUGAAAGCGGGCGUU-UGGCUGCUGGUUAAUGAUGAUACCGGAUAACCGGCACUUAAAGGAUAUUUAUGCUACA--AGUGUAC--- -----------------------.(((((((....((((....)))-).))))))).............((((....))))(((((..((.((....)).))..)--))))...--- ( -24.00) >DroPse_CAF1 101783 111 + 1 UUCCAUCCUGCGACAGGGGAACGGCAGUAGCGUGAAAGCGAGCGUU-UGCCU-CCGGUUAAUGAUGAUACCGGAUAACCAGCACUUAAAGGAUAUUUAUGCUGCAAUGUGAG----C ((((..((((...)))))))).(((((...(((....))).....)-))))(-(((((..........))))))....(((((..((((.....))))))))).........----. ( -29.70) >DroGri_CAF1 87224 88 + 1 -----------------------GUAGCAGCGUGAAAGCGGGCGUU-UGGCUGCUGGUUAAUGAUGAUACCGGAUAACCGGCACUUAAAGGAUAUUUAUGCUGUA--AGUAUAC--- -----------------------...((((((((((......(.((-(((.(((((((((.((.......))..))))))))).))))).)...)))))))))).--.......--- ( -26.90) >DroMoj_CAF1 102147 91 + 1 -----------------------GCAGCAGCGUGAAAGCAGGCGUUUUGGCUGCUGGUUAAUGAUGAUACCAGAUAACCGGCACUUAAAGGAUAUUUAUGCUGCAAAAGUGAAC--- -----------------------((((((((.((....)).)).((((((.(((((((((.((.......))..))))))))).))))))........))))))..........--- ( -29.50) >DroAna_CAF1 84310 95 + 1 ---GGUU-------------GGUGCUGUAGCGUGAAAGCGGGCGUU-UGCCU-CCGGUUAAUGAUGAUACCGGAUAACCGGCACUUAAAGGAUAUUUAUGCUGCAAUGCCAG----C ---.(((-------------(((..(((((((((((...((((...-.))))-(((((((.((.......))..))))))).............)))))))))))..)))))----) ( -35.60) >consensus _______________________GCAGCAGCGUGAAAGCGGGCGUU_UGCCU_CCGGUUAAUGAUGAUACCGGAUAACCGGCACUUAAAGGAUAUUUAUGCUGCAAUAGUAAAC___ ..........................((((((((((.((.((.(......)..(((((..........)))))....)).)).((....))...))))))))))............. (-20.38 = -20.02 + -0.36)
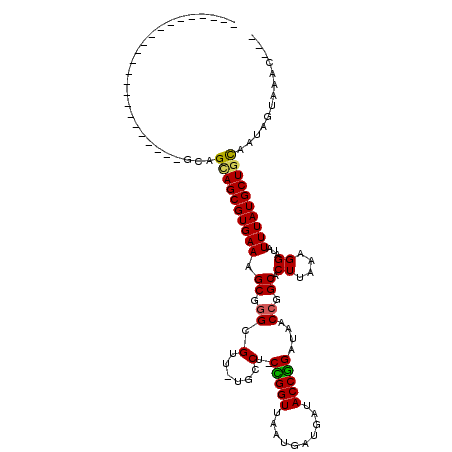
| Location | 4,650,411 – 4,650,508 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.09 |
| Mean single sequence MFE | -23.46 |
| Consensus MFE | -13.83 |
| Energy contribution | -13.97 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4650411 97 - 22407834 CGCGAUCGGCAUUGCAGCAUAAAUAUCCUUUAAGUGCCGGUUAUCCGGUAUCAUCAUUAACCGG-AGGCA-AACGCCCGCUUUCACGCUGCCUCA---------------GAUC--- ...((((......(((((...............(((((((....)))))))...........((-((((.-.......))))))..)))))....---------------))))--- ( -28.70) >DroVir_CAF1 90903 88 - 1 ---GUACACU--UGUAGCAUAAAUAUCCUUUAAGUGCCGGUUAUCCGGUAUCAUCAUUAACCAGCAGCCA-AACGCCCGCUUUCACGCUGCUGC----------------------- ---...((((--((.((.((....)).)).))))))((((....)))).............(((((((..-...............))))))).----------------------- ( -19.93) >DroPse_CAF1 101783 111 - 1 G----CUCACAUUGCAGCAUAAAUAUCCUUUAAGUGCUGGUUAUCCGGUAUCAUCAUUAACCGG-AGGCA-AACGCUCGCUUUCACGCUACUGCCGUUCCCCUGUCGCAGGAUGGAA .----.........((((((.............))))))....((((((..........)))))-)((((-...((..........))...)))).((((((((...))))..)))) ( -26.12) >DroGri_CAF1 87224 88 - 1 ---GUAUACU--UACAGCAUAAAUAUCCUUUAAGUGCCGGUUAUCCGGUAUCAUCAUUAACCAGCAGCCA-AACGCCCGCUUUCACGCUGCUAC----------------------- ---.......--..((((...............(((((((....)))))))...........(((.((..-...))..))).....))))....----------------------- ( -18.00) >DroMoj_CAF1 102147 91 - 1 ---GUUCACUUUUGCAGCAUAAAUAUCCUUUAAGUGCCGGUUAUCUGGUAUCAUCAUUAACCAGCAGCCAAAACGCCUGCUUUCACGCUGCUGC----------------------- ---..........(((((...............(((((((....)))))))...........(((((.(.....).))))).....)))))...----------------------- ( -22.20) >DroAna_CAF1 84310 95 - 1 G----CUGGCAUUGCAGCAUAAAUAUCCUUUAAGUGCCGGUUAUCCGGUAUCAUCAUUAACCGG-AGGCA-AACGCCCGCUUUCACGCUACAGCACC-------------AACC--- (----(((((...))(((...............(((((((....)))))))...........((-((((.-.......))))))..))).))))...-------------....--- ( -25.80) >consensus ___GUACACCAUUGCAGCAUAAAUAUCCUUUAAGUGCCGGUUAUCCGGUAUCAUCAUUAACCAG_AGCCA_AACGCCCGCUUUCACGCUGCUGC_______________________ .............(((((.((((.....)))).(((((((....)))))))...................................))))).......................... (-13.83 = -13.97 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:53 2006