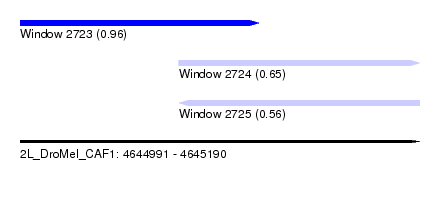
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,644,991 – 4,645,190 |
| Length | 199 |
| Max. P | 0.957544 |

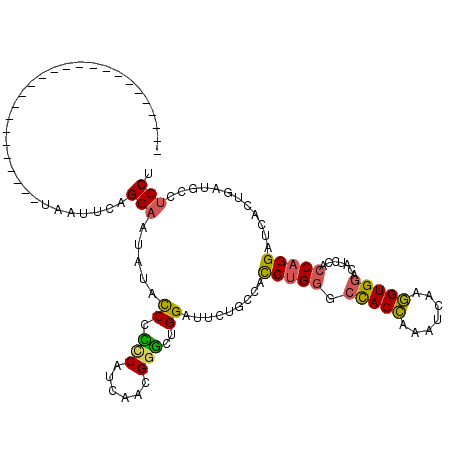
| Location | 4,644,991 – 4,645,110 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 65.89 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -13.16 |
| Energy contribution | -12.85 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4644991 119 + 22407834 AUCCACCAGGAUCACAGAAGCCUCCUAAUGUGGGAAUAUACCCCCCAUCCACGGGCUGGAUUCCGCCAUCUGGGCCACUAACUCAAAGUGGACAUCCACCAGGAUCACUGUUGCCUCCU ...((.(((((((.....(((((....(((.(((........))))))....)))))(((((((((....((((.......))))..))))).)))).....)))).))).))...... ( -33.70) >DroSec_CAF1 81813 119 + 1 AUCCACCAGGAUCACAGAAGCCUCCUAAUUCCGGUAUAUACCCCCCAUCCACGGGCUGGACUUUGCCAUCUGGGCUACCAAAUCAAGGUGGAAAUCCACCAGGAUCAAUGGUGCCUCCA ........(((...(((((((((.........(((....)))..........)))))((......)).))))(((.((((.(((..(((((....)))))..)))...)))))))))). ( -36.21) >DroEre_CAF1 86018 94 + 1 -------------------------UAAUUCUGGAUUACAUCCUCCACAACAGAACUGGAUUCUGCCUCCUGGACCACCAAACCAAGGUGAACAUCCACCUGGAUCACUGAUGCCUCCC -------------------------.......(((...((((.((((...(((((.....))))).....))))........(((.((((......)))))))......))))..))). ( -20.20) >DroYak_CAF1 85761 94 + 1 -------------------------UAACUUAGGAACAAACCCUUCAUUACCGGGCUGGAUUCUGCCUCCUGCGCCACCAAAUCUAGGUGGACAUCCACCAGGAACACAGCUGCCUCCU -------------------------......((((......((.........))((((..((((((.....)).............(((((....))))).))))..))))....)))) ( -23.00) >consensus _________________________UAAUUCAGGAAUAUACCCCCCAUCAACGGGCUGGAUUCUGCCACCUGGGCCACCAAAUCAAGGUGGACAUCCACCAGGAUCACUGAUGCCUCCU ................................(((.....((.(((......)))..)).........(((((.(((((.......))))).......)))))............))). (-13.16 = -12.85 + -0.31)

| Location | 4,645,070 – 4,645,190 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -28.13 |
| Energy contribution | -29.47 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4645070 120 + 22407834 AACUCAAAGUGGACAUCCACCAGGAUCACUGUUGCCUCCUAAUACUGGAUUGCCACCGGGCUCUAUUCCGCCUCUUCGACCACCAAAUCAAGGUGGACAUCCACCAGGAUCACAGAAGCC ...((...((((....))))...))...((((....((((.....(((((..(((((((((........))))....((........))..)))))..)))))..))))..))))..... ( -30.90) >DroSec_CAF1 81892 120 + 1 AAAUCAAGGUGGAAAUCCACCAGGAUCAAUGGUGCCUCCAAUUUCUGUAUUGCCACCGGGCUCUAUUCCGCCUCUUCGACCACCAAUUCAGGGUGGACAUCCACCAGGAUCACUGAAGCC ..(((..((((((..((((((..((....(((((....((((......)))).....((((........)))).......)))))..))..))))))..))))))..))).......... ( -37.10) >DroSim_CAF1 74192 120 + 1 AAAUCAAGGUGGAAAUCCACCAGGAUCAAUGGUGCCUCCAAUUUCUGUAUUGCCACCGGGCUCUAUUCCGCCUCUUCGACCACCAAUUCAAGGUGGACAACCACCAGGAUCACAGAAGCC ..(((..(((((....)))))..)))...(((.....))).(((((((.((((((((((((........))))....((........))..))))).)))((....))...))))))).. ( -34.30) >consensus AAAUCAAGGUGGAAAUCCACCAGGAUCAAUGGUGCCUCCAAUUUCUGUAUUGCCACCGGGCUCUAUUCCGCCUCUUCGACCACCAAUUCAAGGUGGACAUCCACCAGGAUCACAGAAGCC ......((((((((...(((((.......))))).................(((....)))....))))))))((((..(((((.......)))))..((((....))))....)))).. (-28.13 = -29.47 + 1.33)


| Location | 4,645,070 – 4,645,190 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -43.57 |
| Consensus MFE | -37.45 |
| Energy contribution | -38.23 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4645070 120 - 22407834 GGCUUCUGUGAUCCUGGUGGAUGUCCACCUUGAUUUGGUGGUCGAAGAGGCGGAAUAGAGCCCGGUGGCAAUCCAGUAUUAGGAGGCAACAGUGAUCCUGGUGGAUGUCCACUUUGAGUU ...........((..((((((((((((((.....((((..((((..(.(((........))))..))))...))))....((((.((....))..))))))))))))))))))..))... ( -44.80) >DroSec_CAF1 81892 120 - 1 GGCUUCAGUGAUCCUGGUGGAUGUCCACCCUGAAUUGGUGGUCGAAGAGGCGGAAUAGAGCCCGGUGGCAAUACAGAAAUUGGAGGCACCAUUGAUCCUGGUGGAUUUCCACCUUGAUUU .........((((..((((((.(((((((..(.((..(((((((..(.(((........))))..))((....(((...)))...)))))))..)).).))))))).))))))..)))). ( -43.20) >DroSim_CAF1 74192 120 - 1 GGCUUCUGUGAUCCUGGUGGUUGUCCACCUUGAAUUGGUGGUCGAAGAGGCGGAAUAGAGCCCGGUGGCAAUACAGAAAUUGGAGGCACCAUUGAUCCUGGUGGAUUUCCACCUUGAUUU .........((((..(((((..(((((((..((.(..(((((((..(.(((........))))..))((....(((...)))...)))))))..)))..)))))))..)))))..)))). ( -42.70) >consensus GGCUUCUGUGAUCCUGGUGGAUGUCCACCUUGAAUUGGUGGUCGAAGAGGCGGAAUAGAGCCCGGUGGCAAUACAGAAAUUGGAGGCACCAUUGAUCCUGGUGGAUUUCCACCUUGAUUU .........((((..((((((.(((((((..((..(((((((((..(.(((........))))..))))....(.......)....)))))....))..))))))).))))))..)))). (-37.45 = -38.23 + 0.78)

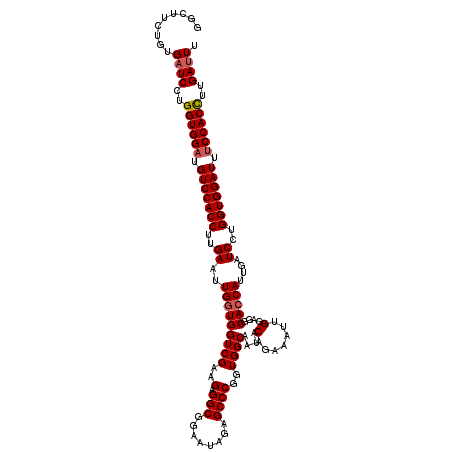
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:52 2006