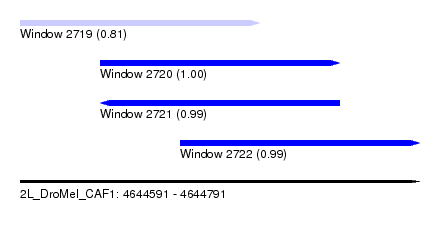
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,644,591 – 4,644,791 |
| Length | 200 |
| Max. P | 0.997711 |

| Location | 4,644,591 – 4,644,711 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.72 |
| Mean single sequence MFE | -33.91 |
| Consensus MFE | -15.10 |
| Energy contribution | -16.10 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4644591 120 + 22407834 CCACCGGGUUCCAUUCCGCCUCUUCGACCACCAAAUCAAGGUGGACAUCCACCAGGAUCACAGAAGCCUCCUAAUGUGGGAAUAUACCCCCCAUCCACGGGCUGGAUUCCGCCAUUUGGG (((.(((..((((..(((...((((..(((((.......)))))..((((....))))....)))).......(((.(((........))))))...)))..))))..))).....))). ( -36.30) >DroSec_CAF1 81413 120 + 1 CCACCAGGAUCUAUGCCGCCUCUUGGAUCACCGAAUCAAAUUGGAUAUCCACCAGGAUCACUGAAGCCUCCUAAUUCCGGUAUAUAUCCCCCAUCAACGGGCUGGACUUUGCCAUCUGGU ..((((((((.((((((((((..(((((..((((......))))..)))))..)))......(((.........)))))))))).))))..........(((........)))...)))) ( -32.20) >DroEre_CAF1 85779 102 + 1 ------------------CCUCCUGGGCCACCAAACCAAGGUGGAUAUCCACCAGGAUCACAGAUGCCUCCUAAUUCUGGAUUACAUCCUCCACAACAGAAUUGGAUUCUGCCUCCUGGG ------------------.....(((((((((.......)))))...))))((((((...((((....(((.(((((((.................)))))))))).))))..)))))). ( -33.23) >consensus CCACC_GG_UC_AU_CCGCCUCUUGGACCACCAAAUCAAGGUGGAUAUCCACCAGGAUCACAGAAGCCUCCUAAUUCCGGAAUAUAUCCCCCAUCAACGGGCUGGAUUCUGCCAUCUGGG ..............................(((......(((((....)))))((((...........))))......((.....(((((((......)))..))))....))...))). (-15.10 = -16.10 + 1.00)

| Location | 4,644,631 – 4,644,751 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -40.54 |
| Consensus MFE | -26.83 |
| Energy contribution | -27.83 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4644631 120 + 22407834 GUGGACAUCCACCAGGAUCACAGAAGCCUCCUAAUGUGGGAAUAUACCCCCCAUCCACGGGCUGGAUUCCGCCAUUUGGGCCACUAACUCAAGGUGGACAUCCACCAGGAUCACUGUUGC ((((....))))(((((((.....(((((....(((.(((........))))))....)))))((((((((((..(((((.......))))))))))).)))).....)))).))).... ( -42.40) >DroSec_CAF1 81453 120 + 1 UUGGAUAUCCACCAGGAUCACUGAAGCCUCCUAAUUCCGGUAUAUAUCCCCCAUCAACGGGCUGGACUUUGCCAUCUGGUCUACCAAAUCAAGGUGGAAAUCCACCAGGAUCAAUGGUGC ..((((.((((((.(((.....((....)).....)))((((....((((((......)))..)))...))))...(((....)))......)))))).))))((((.......)))).. ( -35.80) >DroEre_CAF1 85801 120 + 1 GUGGAUAUCCACCAGGAUCACAGAUGCCUCCUAAUUCUGGAUUACAUCCUCCACAACAGAAUUGGAUUCUGCCUCCUGGGCCACCAAACCAAGGUGGAUAUCCACCAGGAUCACAGAUGC (((((((((((((((((...((((....(((.(((((((.................)))))))))).))))..))))((....)).......)))))))))))))............... ( -43.43) >consensus GUGGAUAUCCACCAGGAUCACAGAAGCCUCCUAAUUCCGGAAUAUAUCCCCCAUCAACGGGCUGGAUUCUGCCAUCUGGGCCACCAAAUCAAGGUGGAAAUCCACCAGGAUCACUGAUGC ((((((.((((((((((...........))))......((.....(((((((......)))..))))....))...(((....)))......)))))).))))))............... (-26.83 = -27.83 + 1.00)


| Location | 4,644,631 – 4,644,751 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -45.20 |
| Consensus MFE | -36.17 |
| Energy contribution | -35.63 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4644631 120 - 22407834 GCAACAGUGAUCCUGGUGGAUGUCCACCUUGAGUUAGUGGCCCAAAUGGCGGAAUCCAGCCCGUGGAUGGGGGGUAUAUUCCCACAUUAGGAGGCUUCUGUGAUCCUGGUGGAUGUCCAC ....(((.....)))(((((((((((((..((..(((.((((((...(((........)))..))((((.((((.....)))).))))....)))).)))...))..))))))))))))) ( -47.80) >DroSec_CAF1 81453 120 - 1 GCACCAUUGAUCCUGGUGGAUUUCCACCUUGAUUUGGUAGACCAGAUGGCAAAGUCCAGCCCGUUGAUGGGGGAUAUAUACCGGAAUUAGGAGGCUUCAGUGAUCCUGGUGGAUAUCCAA ...((((..((...(((((....)))))...((((((....))))))(((........))).))..)))).((((((.((((((.((((.(......)..)))).)))))).)))))).. ( -41.40) >DroEre_CAF1 85801 120 - 1 GCAUCUGUGAUCCUGGUGGAUAUCCACCUUGGUUUGGUGGCCCAGGAGGCAGAAUCCAAUUCUGUUGUGGAGGAUGUAAUCCAGAAUUAGGAGGCAUCUGUGAUCCUGGUGGAUAUCCAC ...............(((((((((((((..((((....)))).((((.(((((..((..(((((((.((((........)))).)).)))))))..)))))..))))))))))))))))) ( -46.40) >consensus GCAACAGUGAUCCUGGUGGAUAUCCACCUUGAUUUGGUGGCCCAGAUGGCAGAAUCCAGCCCGUUGAUGGGGGAUAUAUUCCAGAAUUAGGAGGCUUCUGUGAUCCUGGUGGAUAUCCAC ...............(((((((((((((....(((((..(((.....)))...((((..(((......))))))).....)))))...((((.((....))..))))))))))))))))) (-36.17 = -35.63 + -0.54)

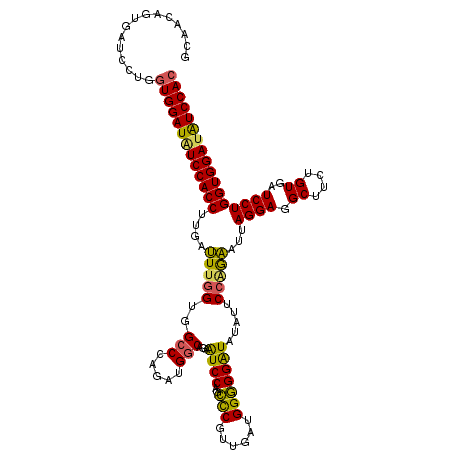
| Location | 4,644,671 – 4,644,791 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.19 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -14.77 |
| Energy contribution | -15.14 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4644671 120 + 22407834 AAUAUACCCCCCAUCCACGGGCUGGAUUCCGCCAUUUGGGCCACUAACUCAAGGUGGACAUCCACCAGGAUCACUGUUGCCUCCUAAUACUGGAUUGCCACCGGGCUCUAUUCCGCCGCU ............((((...((.(((((((((((..(((((.......))))))))))).))))))).))))..(.((.((.(((.......)))..)).)).)(((........)))... ( -33.10) >DroSec_CAF1 81493 120 + 1 UAUAUAUCCCCCAUCAACGGGCUGGACUUUGCCAUCUGGUCUACCAAAUCAAGGUGGAAAUCCACCAGGAUCAAUGGUGCCUCCAAUUUCUGUAUUGCCACCGGGCUCUAUUCCGCCUCU ..................((((.(((....(((...(((..(((((.(((..(((((....)))))..)))...)))))....((((......)))))))...))).....))))))).. ( -32.20) >DroEre_CAF1 85841 95 + 1 AUUACAUCCUCCACAACAGAAUUGGAUUCUGCCUCCUGGGCCACCAAACCAAGGUGGAUAUCCACCAGGAUCACAGAUGCCUCCUAAUUCUGGAU------------------------- ................((((((((((.((((..((((((.(((((.......))))).......))))))...))))....))).)))))))...------------------------- ( -31.50) >DroYak_CAF1 85584 95 + 1 AUUAAACCCUCCACUACCGGGCUGGAUUCUGCCUCCUGCGCCACCAAAUCUAGGUGGACAUCCACCAGGAACACAGCUGCCUCCUAGUUCAGGAU------------------------- .......(((..((((..(((((((....(((.....)))...)))..(((.(((((....))))).)))........))))..))))..)))..------------------------- ( -26.30) >consensus AAUAUACCCCCCACCAACGGGCUGGAUUCUGCCACCUGGGCCACCAAAUCAAGGUGGACAUCCACCAGGAUCACAGAUGCCUCCUAAUUCUGGAU_________________________ ..................((((.........((...(((....)))......(((((....))))).)).........))))...................................... (-14.77 = -15.14 + 0.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:49 2006