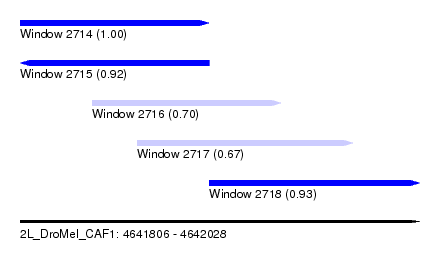
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,641,806 – 4,642,028 |
| Length | 222 |
| Max. P | 0.999297 |

| Location | 4,641,806 – 4,641,911 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.86 |
| Mean single sequence MFE | -34.35 |
| Consensus MFE | -18.93 |
| Energy contribution | -20.27 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.55 |
| SVM decision value | 3.49 |
| SVM RNA-class probability | 0.999297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4641806 105 + 22407834 AAUUUCCAGGAUUACUGGGAUCAACGGGCUCGGGUCCAUCGGGAAUACCAAAUCUUCCCGG------------GACAAAUGCCUCUCCAGGUUGGAGUUCACCUGGAUCUUCGAUUC .....((((.....))))((((...((((....((((..((((((.........)))))))------------)))....)))).(((((((........))))))).....)))). ( -37.50) >DroSec_CAF1 78834 105 + 1 AACCUCCAGGAUUAAUAGGACCAACAGUCUCGAGUCCAUCGGGAAUUCCAAAUCUUCCCGG------------GACACACGUCUCUCCAGGUUGGAGUUCUCCUGGAUCUUCGAUUC ....(((((((......(((((.........).)))).....(((((((((.(((....((------------(((....)))))...))))))))))))))))))).......... ( -32.90) >DroYak_CAF1 83719 117 + 1 AACCUCCAGGAAUACAAGGACCACCUGGCUCCAUUGCAUUGGGACCACCAUAUCCUCCCCAACAAUCUCCUAUAACAUCUGCAACGCCAGGUUGGAGUUCUCCUGGAAUUUCGAUUC ....(((((((..((.....(((((((((....((((((((((..............))))).................))))).)))))).))).))..))))))).......... ( -32.66) >consensus AACCUCCAGGAUUACUAGGACCAACAGGCUCGAGUCCAUCGGGAAUACCAAAUCUUCCCGG____________GACAAAUGCCUCUCCAGGUUGGAGUUCUCCUGGAUCUUCGAUUC ....((((((.......((((............)))).(((((((.........))))))).......................((((.....))))....)))))).......... (-18.93 = -20.27 + 1.34)

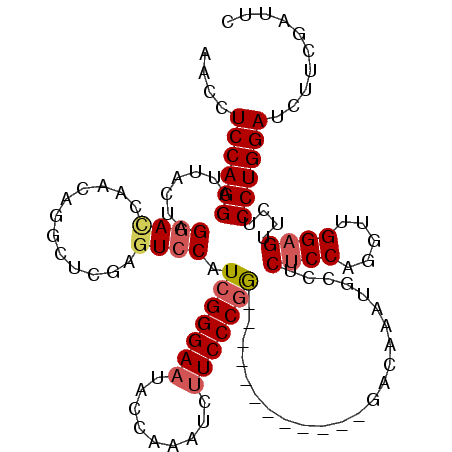

| Location | 4,641,806 – 4,641,911 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.86 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -21.35 |
| Energy contribution | -22.47 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4641806 105 - 22407834 GAAUCGAAGAUCCAGGUGAACUCCAACCUGGAGAGGCAUUUGUC------------CCGGGAAGAUUUGGUAUUCCCGAUGGACCCGAGCCCGUUGAUCCCAGUAAUCCUGGAAAUU ..(((((...(((((((........)))))))(.(((....(((------------(((((((.........))))))..))))....)))).))))).((((.....))))..... ( -36.00) >DroSec_CAF1 78834 105 - 1 GAAUCGAAGAUCCAGGAGAACUCCAACCUGGAGAGACGUGUGUC------------CCGGGAAGAUUUGGAAUUCCCGAUGGACUCGAGACUGUUGGUCCUAUUAAUCCUGGAGGUU ..........(((((((...((((.....))))...((...(((------------(((((((.........))))))..)))).)).(((.....))).......))))))).... ( -34.00) >DroYak_CAF1 83719 117 - 1 GAAUCGAAAUUCCAGGAGAACUCCAACCUGGCGUUGCAGAUGUUAUAGGAGAUUGUUGGGGAGGAUAUGGUGGUCCCAAUGCAAUGGAGCCAGGUGGUCCUUGUAUUCCUGGAGGUU .........(((((((((.((.(((.(((((((((((.(((.((....)).)))(((((((..(......)..))))))))))))...))))))))).....)).)))))))))... ( -41.00) >consensus GAAUCGAAGAUCCAGGAGAACUCCAACCUGGAGAGGCAUAUGUC____________CCGGGAAGAUUUGGUAUUCCCGAUGGACUCGAGCCAGUUGGUCCUAGUAAUCCUGGAGGUU ..........(((((((...((((.....)))).........................((((......((....))((((((........)))))).)))).....))))))).... (-21.35 = -22.47 + 1.12)

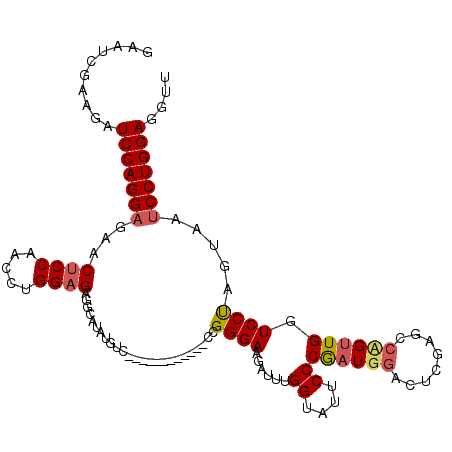
| Location | 4,641,846 – 4,641,951 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 63.88 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -11.10 |
| Energy contribution | -14.16 |
| Covariance contribution | 3.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4641846 105 + 22407834 GGGAAUACCAAAU---CUUCCCGG------------GACAAAUGCCUCUCCAGGUUGGAGUUCACCUGGAUCUUCGAUUCCUUCCUACCAACUGCCAGGAAUACAACAGCAACCAGGCUC (((((........---.)))))..------------.......((((.(((((((........)))))))......((((((..(........)..))))))............)))).. ( -25.80) >DroSec_CAF1 78874 105 + 1 GGGAAUUCCAAAU---CUUCCCGG------------GACACACGUCUCUCCAGGUUGGAGUUCUCCUGGAUCUUCGAUUCCUUCCGACCAACUUCCAGGAAUACAACUGCAACCAGGCUC (((((........---.)))))((------------(((....))))).((.(((((.((((.(((((((...(((........)))......)))))))....)))).))))).))... ( -35.50) >DroEre_CAF1 84000 96 + 1 --------UAUGCCUGCCUCUCCACCC--------------AUACA--ACCAGGUUGGAGUCCACCUGGAUCACCUUCUCCUCCCUUCCCACCGCCAGGACUACCGGAAACACCAGGUUG --------...(((((....(((....--------------.....--.((((((.((...)))))))).........((((..(........)..)))).....))).....))))).. ( -21.60) >DroYak_CAF1 83759 117 + 1 GGGACCACCAUAU---CCUCCCCAACAAUCUCCUAUAACAUCUGCAACGCCAGGUUGGAGUUCUCCUGGAAUUUCGAUUCCUUCCUUCCAACACCCAGGGCUACAACAGCCACCAGGCGC ((((.........---..)))).........................((((..(((((((.......(((((....)))))....)))))))......((((.....))))....)))). ( -27.60) >consensus GGGAAUACCAAAU___CCUCCCCA____________GACA_AUGCAUCUCCAGGUUGGAGUUCACCUGGAUCUUCGAUUCCUUCCUACCAACUGCCAGGAAUACAACAGCAACCAGGCUC (((((............)))))..........................(((.(((((..((..((((((.........................))))))..)).....))))).))).. (-11.10 = -14.16 + 3.06)

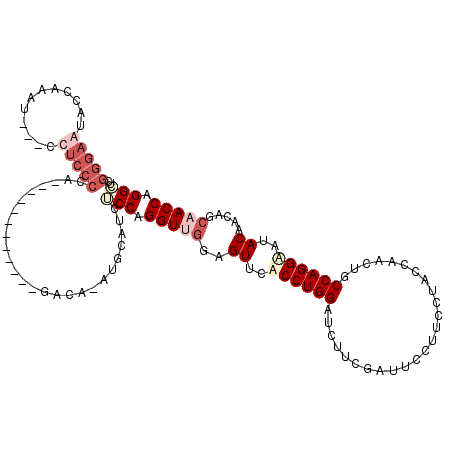
| Location | 4,641,871 – 4,641,991 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 63.33 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -13.53 |
| Energy contribution | -13.54 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4641871 120 + 22407834 AAUGCCUCUCCAGGUUGGAGUUCACCUGGAUCUUCGAUUCCUUCCUACCAACUGCCAGGAAUACAACAGCAACCAGGCUCAAGUCCAGUAGAAUAUCCACAUGCUCCUAUCAAACCUCCA ...........((((((((((.....(((((.(((.((((((..(........)..)))))).........((..(((....)))..)).))).)))))...))))).....)))))... ( -24.60) >DroSec_CAF1 78899 104 + 1 CACGUCUCUCCAGGUUGGAGUUCUCCUGGAUCUUCGAUUCCUUCCGACCAACUUCCAGGAAUACAACUGCAACCAGGCUCAAGUUCUG------GUCCUCAAG----------UCCUCCA ............(((((.((((.(((((((...(((........)))......)))))))....)))).)))))(((..((.....))------..)))....----------....... ( -26.40) >DroEre_CAF1 84019 117 + 1 -AUACA--ACCAGGUUGGAGUCCACCUGGAUCACCUUCUCCUCCCUUCCCACCGCCAGGACUACCGGAAACACCAGGUUGGAAUCCAUGGGGGUCUGAGAAUCGUCCCAUCCAUCCUCCG -.....--...(((.((((((((....))))....((((((((((((((.(((....((....))(....)....))).)))).....)))))...)))))........)))).)))... ( -32.60) >consensus _AUGCCUCUCCAGGUUGGAGUUCACCUGGAUCUUCGAUUCCUUCCUACCAACUGCCAGGAAUACAACAGCAACCAGGCUCAAGUCCAG__G__UAUCCACAUG_UCC_AUC_AUCCUCCA ................(((.((..(((((.......((((((..............))))))..........)))))...)).))).................................. (-13.53 = -13.54 + 0.00)


| Location | 4,641,911 – 4,642,028 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 57.42 |
| Mean single sequence MFE | -30.51 |
| Consensus MFE | -12.58 |
| Energy contribution | -11.81 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4641911 117 + 22407834 CUUCCUACCAACUGCCAGGAAUACAACAGCAACCAGGCUCAAGUCCAGUAGAAUAUCCACAUGCUCCUAUCAAACCUCCAA---GUUCCGCCCCGGCAGGUUGGAGCUCACCAGGAUCAC ..((((.....((((..(((.......(((......)))....))).))))...........(((((.....(((((((..---..........)).)))))))))).....)))).... ( -25.00) >DroSec_CAF1 78939 93 + 1 CUUCCGACCAACUUCCAGGAAUACAACUGCAACCAGGCUCAAGUUCUG------GUCCUCAAG----------UCCUCCAA---CCUC--------AAGGUUGGAGUCCACCUGGAACUU ............(((((((............(((((((....).))))------))......(----------..((((((---((..--------..))))))))..).)))))))... ( -32.40) >DroEre_CAF1 84056 120 + 1 CUCCCUUCCCACCGCCAGGACUACCGGAAACACCAGGUUGGAAUCCAUGGGGGUCUGAGAAUCGUCCCAUCCAUCCUCCGGCGCAUCCCGUACCACCCGGUUGGAGUUCACCUGGAUCAC ...........((....))......(....).((((((.(((.(((((((((((.((.((..((((.............))))..)).)).))).))))..)))).)))))))))..... ( -34.12) >consensus CUUCCUACCAACUGCCAGGAAUACAACAGCAACCAGGCUCAAGUCCAG__G__UAUCCACAUG_UCC_AUC_AUCCUCCAA___AUUCCG__CC__CAGGUUGGAGUUCACCUGGAUCAC ..............(((((................(((....)))..............................((((((...................))))))....)))))..... (-12.58 = -11.81 + -0.77)

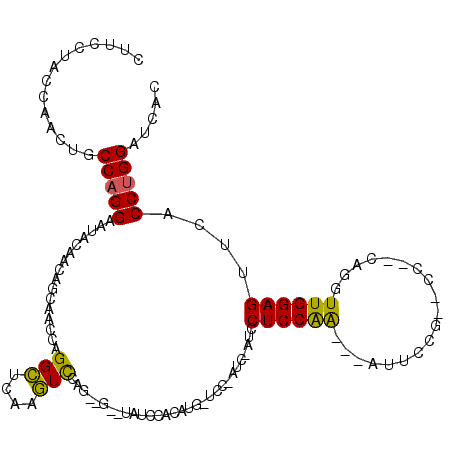

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:45 2006