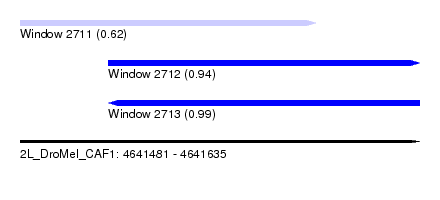
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,641,481 – 4,641,635 |
| Length | 154 |
| Max. P | 0.985520 |

| Location | 4,641,481 – 4,641,595 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.03 |
| Mean single sequence MFE | -20.30 |
| Consensus MFE | -12.35 |
| Energy contribution | -13.23 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4641481 114 + 22407834 CCGUGUAACUCCCCAC------AUCCACUAACCCAAUUUUCCCAAAAGUAUCCAAACUCAGGAUACCUUUGUUAUCCUCAAAAUGAAAUUAAUAAAUACCUAUGGUACCCACCGUGGAGC ..(((........)))------.(((((..............((((.((((((.......)))))).))))................................(((....)))))))).. ( -20.70) >DroSec_CAF1 78512 114 + 1 CCGUGUAAUCCCCCAC------AUCCAAUAAGCCAAUAUUCCCAAAAGUAUCCAAGCCCAGGAUACCUUUGUUAUCCCCAAAAUGAAAUUAAUAAAUACCUGUGGUACCCACCGUGGAGC ............((((------....................((((.((((((.......)))))).))))..............................((((...)))).))))... ( -18.70) >DroEre_CAF1 83704 120 + 1 CCGUGUUAUUAUACAGUAUCAAGCCACAUAAACCAACAUUUCCGAAAGUAUCCAAACACAGGAUACCUGUGUUAUCCACAAAAUGAAAUUAACUCAUACCUCUGGUACCCACCGUGGAGC ..((((....))))............................(....)......((((((((...)))))))).(((((...((((.......))))......(((....)))))))).. ( -22.60) >DroYak_CAF1 83385 120 + 1 CCGUGUUAUAAUCAUGUUUCAAACUACUUAACCCAACAAGCCCAGAACUAUCCAAACCCAGGAUUCCUUUGCUAUCCACAAAAUGAUAUUAAUAGUUAUCUCUGGUACCCACCGUGGGGU .((((.......))))..............((((....(((..((....((((.......))))..))..)))...(((.....((((........))))...(((....)))))))))) ( -19.20) >consensus CCGUGUAAUUACCCAC______ACCAAAUAACCCAACAUUCCCAAAAGUAUCCAAACCCAGGAUACCUUUGUUAUCCACAAAAUGAAAUUAAUAAAUACCUCUGGUACCCACCGUGGAGC ..........................................((((.((((((.......)))))).))))............................(((((((....)))))))... (-12.35 = -13.23 + 0.87)


| Location | 4,641,515 – 4,641,635 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.72 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -17.50 |
| Energy contribution | -18.00 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4641515 120 + 22407834 CCCAAAAGUAUCCAAACUCAGGAUACCUUUGUUAUCCUCAAAAUGAAAUUAAUAAAUACCUAUGGUACCCACCGUGGAGCCAAUGGGUAGGAUAUCAAGGAUAUCAAACAGGAAAUGGAU .(((...((((((.......))))))(((((.(((((((.....))..........((((((((((.((......)).))).)))))))))))).)))))...............))).. ( -33.00) >DroSec_CAF1 78546 117 + 1 CCCAAAAGUAUCCAAGCCCAGGAUACCUUUGUUAUCCCCAAAAUGAAAUUAAUAAAUACCUGUGGUACCCACCGUGGAGCCAAUGGGUAGUA---CAAGGAUAUCCAACAGGAAAUGGAU ..((((.((((((.......)))))).))))......(((..................((((((.((((((...((....)).)))))).))---).)))...(((....)))..))).. ( -32.10) >DroEre_CAF1 83744 117 + 1 UCCGAAAGUAUCCAAACACAGGAUACCUGUGUUAUCCACAAAAUGAAAUUAACUCAUACCUCUGGUACCCACCGUGGAGCCAAUGGGUAGUU---GAAGAAUAUCCACCAGAAAAUGGGU ((...((.((((((((((((((...)))))))).(((((...((((.......))))......(((....)))))))).....)))))).))---...)).......(((.....))).. ( -31.00) >DroYak_CAF1 83425 108 + 1 CCCAGAACUAUCCAAACCCAGGAUUCCUUUGCUAUCCACAAAAUGAUAUUAAUAGUUAUCUCUGGUACCCACCGUGGGGUCAAUGGGUAUCA---CAAGUAUAUCCAACAG--------- .(((((..............((((.........)))).......((((........)))))))))..(((.....))).....((((((((.---...).)))))))....--------- ( -21.60) >consensus CCCAAAAGUAUCCAAACCCAGGAUACCUUUGUUAUCCACAAAAUGAAAUUAAUAAAUACCUCUGGUACCCACCGUGGAGCCAAUGGGUAGUA___CAAGGAUAUCCAACAGGAAAUGGAU ..((((.((((((.......)))))).)))).........................(((((((((..((......))..)).)))))))............................... (-17.50 = -18.00 + 0.50)


| Location | 4,641,515 – 4,641,635 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.72 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -24.50 |
| Energy contribution | -26.88 |
| Covariance contribution | 2.38 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
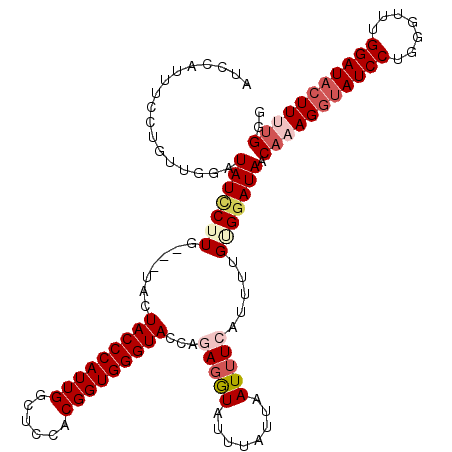
>2L_DroMel_CAF1 4641515 120 - 22407834 AUCCAUUUCCUGUUUGAUAUCCUUGAUAUCCUACCCAUUGGCUCCACGGUGGGUACCAUAGGUAUUUAUUAAUUUCAUUUUGAGGAUAACAAAGGUAUCCUGAGUUUGGAUACUUUUGGG ((((....(((((..((((((...)))))).(((((((((......)))))))))..)))))............((.....))))))..(((((((((((.......))))))))))).. ( -38.00) >DroSec_CAF1 78546 117 - 1 AUCCAUUUCCUGUUGGAUAUCCUUG---UACUACCCAUUGGCUCCACGGUGGGUACCACAGGUAUUUAUUAAUUUCAUUUUGGGGAUAACAAAGGUAUCCUGGGCUUGGAUACUUUUGGG ((((...(((....)))....((((---(..(((((((((......)))))))))..))))).....................))))..(((((((((((.......))))))))))).. ( -39.20) >DroEre_CAF1 83744 117 - 1 ACCCAUUUUCUGGUGGAUAUUCUUC---AACUACCCAUUGGCUCCACGGUGGGUACCAGAGGUAUGAGUUAAUUUCAUUUUGUGGAUAACACAGGUAUCCUGUGUUUGGAUACUUUCGGA ........(((((.((.(((((...---...(((((((((......)))))))))(((.(((.(((((.....)))))))).)))..((((((((...)))))))).))))))).))))) ( -36.60) >DroYak_CAF1 83425 108 - 1 ---------CUGUUGGAUAUACUUG---UGAUACCCAUUGACCCCACGGUGGGUACCAGAGAUAACUAUUAAUAUCAUUUUGUGGAUAGCAAAGGAAUCCUGGGUUUGGAUAGUUCUGGG ---------(((((.((........---...(((((((((......)))))))))((((.(((..((.....((((........))))....))..)))))))..)).)))))....... ( -27.00) >consensus AUCCAUUUCCUGUUGGAUAUCCUUG___UACUACCCAUUGGCUCCACGGUGGGUACCAGAGGUAUUUAUUAAUUUCAUUUUGUGGAUAACAAAGGUAUCCUGGGUUUGGAUACUUUUGGG .................(((((((.......(((((((((......)))))))))...(((((........))))).....))))))).(((((((((((.......))))))))))).. (-24.50 = -26.88 + 2.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:41 2006