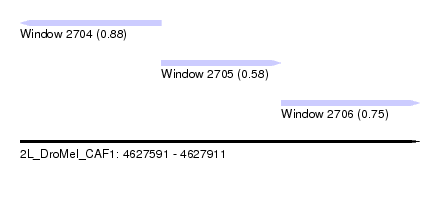
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,627,591 – 4,627,911 |
| Length | 320 |
| Max. P | 0.882901 |

| Location | 4,627,591 – 4,627,704 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 96.81 |
| Mean single sequence MFE | -29.34 |
| Consensus MFE | -28.01 |
| Energy contribution | -28.01 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
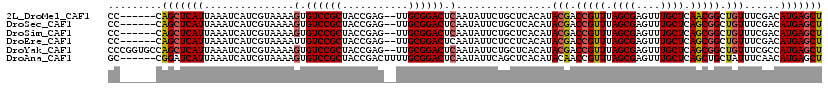
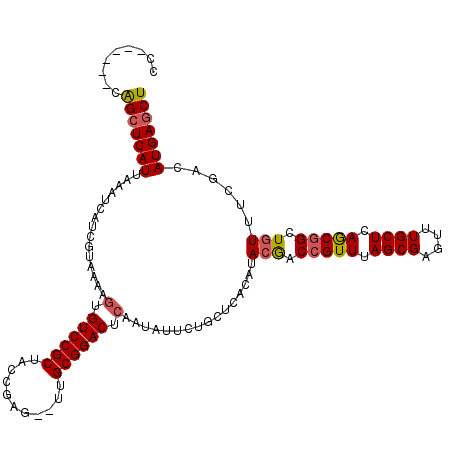
>2L_DroMel_CAF1 4627591 113 - 22407834 UAUUUUAUUGGCGCUCGGGGCGCUGCCGAGAGCACAAAUCAAAACUAACUGGAGUGCUUAGAGUGCCAUAGAGAAACAAACAACAUUAUUCACCGCUCUGAUUUGGUCAAACA ..((((((.(((((((..((((((.(((..((............))...)))))))))..)))))))))))))..................((((........))))...... ( -29.00) >DroSec_CAF1 64858 113 - 1 UAUUUUAUUGGCGCUCGGGGCGCUGCCGAGAGCACAAAUCAAAACUAACUGGAGUGCUUAGAGUGCCAUAGAGAAACAAACAACAUUAUUCACCGCUCUGAUUUGGCAAAACA ..((((((.(((((((..((((((.(((..((............))...)))))))))..))))))))))))).....................(((.......)))...... ( -29.20) >DroSim_CAF1 64010 113 - 1 UAUUUUAUUGGCGCUCGGGGCGCUGCCGAGAGCACAAAUCAAAACUAACUGGAGUGCUUAGAGUGCCAUAGAGAAACAAACAACAUUAUUCCCCGCUCUGAUUUGGCCAAACA .......(((((((((..(((...)))..)))).((((((...(((......)))....((((((............................)))))))))))))))))... ( -28.59) >DroEre_CAF1 69905 113 - 1 UAUUUCAUUGGCGCUCGGGGCGCUGCCGUAAGCACAAAUCAAAACUAACUGGAGUGCUUAGAGUGCCAUAGAGAAACAAACAACAUUAUUUACCGCUCUGAUUUGGUCAAACA ..((((..((((((((..((((((.(.((.((............)).)).).))))))..))))))))..)))).................((((........))))...... ( -30.60) >DroYak_CAF1 69520 113 - 1 UAUUUUAUUGGCGCUCGGGGCGCUGCCGAGAGCACAAAUCAAAUCUAACUGGAGUGCUUAGAGUGCCAUAGAGAAACAAACAACAUUAUUCACCGCUCUGAUUUGGACAAACA ..((((((.(((((((..((((((.((((((............)))...)))))))))..)))))))))))))...................(((........)))....... ( -29.30) >consensus UAUUUUAUUGGCGCUCGGGGCGCUGCCGAGAGCACAAAUCAAAACUAACUGGAGUGCUUAGAGUGCCAUAGAGAAACAAACAACAUUAUUCACCGCUCUGAUUUGGCCAAACA ........((((((((..((((((.(((..((............))...)))))))))..))))))))(((((......................)))))............. (-28.01 = -28.01 + -0.00)

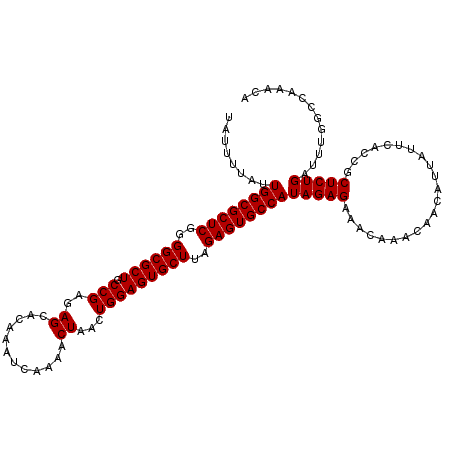

| Location | 4,627,704 – 4,627,800 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 96.47 |
| Mean single sequence MFE | -35.84 |
| Consensus MFE | -33.30 |
| Energy contribution | -33.42 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4627704 96 + 22407834 CAUUUAAGUAGAAACUGUGCCGCUUCUGGUCCGCUCGCCA-ACCCGUGUUACGCGUCGGAGUUUCUGGUGGGGAUCAGACGUGGCCUCAGUCCCGGU ..........(..((((.(((((.((((((((.(.(((((-(((((((.....)).))).))...)))))))))))))).)))))..))))..)... ( -38.60) >DroSec_CAF1 64971 96 + 1 CAUUUAAGUAGAAACUGUGCCGCUUCUGGUCCGCUCGCCA-ACCCGUGUUACGCGUCGGAGUUUCUGUUGGGGAUCGGACGUGGUCUCAGUCCCGGU ..........(..((((.(((((.((((((((.(.((..(-(((((((.....)).))).)))..))..).)))))))).)))))..))))..)... ( -32.30) >DroSim_CAF1 64123 96 + 1 CAUUUAAGUAGAAACUGUGCCGCUUCUGGUCCGCUCGCCA-ACCCGUGUUACGCGUCGGAGUUUCUGUUGGGGAUCGGACGUGGUCACAGUCCCGGU ..........(..((((((((((.((((((((.(.((..(-(((((((.....)).))).)))..))..).)))))))).)))).))))))..)... ( -35.80) >DroEre_CAF1 70018 96 + 1 CAUUUAAGUAAAAACUGUGCCGCUUCUGGUCCGCUCGCCA-ACCCGUGUUACGCGUCGGAGUUUCUGGCGGGGAUCGGACGUGGUCUCAGUCCCGGU .............((((.(((((.((((((((.(.(((((-(((((((.....)).))).))...)))))))))))))).)))))..))))...... ( -37.40) >DroYak_CAF1 69633 97 + 1 CAUUUAAGUAGAAACUGUGCCGCUUCUGGUCCGCUCGCCAGACCCGUGUUACGCGUCGGAGUUUCUGGUGGAGAUCGGACGUGGUCUCAGUCCCGGU ..........(..((((.(((((.(((((((...((((((((((((((.....)).))).)..)))))))).))))))).)))))..))))..)... ( -35.10) >consensus CAUUUAAGUAGAAACUGUGCCGCUUCUGGUCCGCUCGCCA_ACCCGUGUUACGCGUCGGAGUUUCUGGUGGGGAUCGGACGUGGUCUCAGUCCCGGU .............((((.(((((.((((((((.(.(((((.(((((((.....)).))).))...)))))))))))))).)))))..))))...... (-33.30 = -33.42 + 0.12)

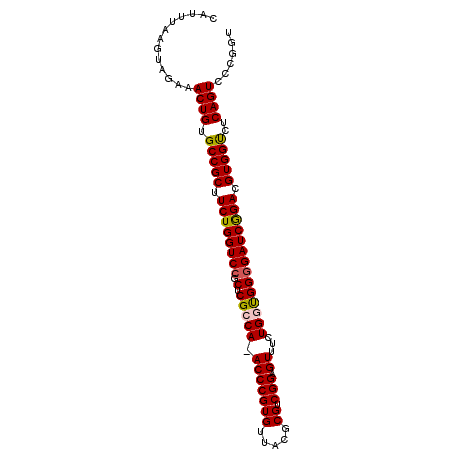
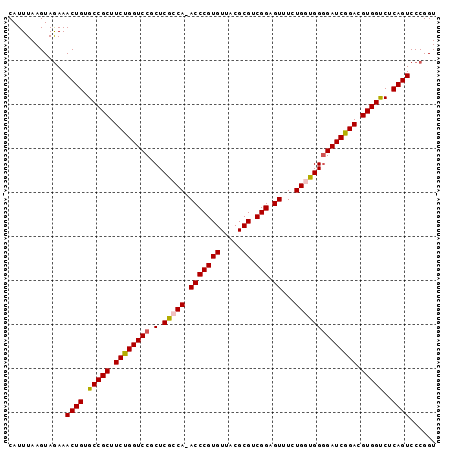
| Location | 4,627,800 – 4,627,911 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.84 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -27.25 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4627800 111 + 22407834 CC------CAGCUCAUUAAAUCAUCGUAAAAGUGUCCGCUACCGAG--UUGCGGACUCAAUAUUCUGCUCACAUACGACCGUUUAGCGAGUUUGCUCAACGGCUGUUUCGACAUGAGCU ..------.(((((((.......(((((...(.((((((.......--..)))))).).......((....)))))))(((((.((((....)))).)))))..........))))))) ( -31.80) >DroSec_CAF1 65067 111 + 1 CC------CAGCUCAUUAAAUCAUCGUAAAAGUGUCCGCUACCGAG--UUGCGGACUCAAUAUUCUGCUCACAUACGACCGUUUAGCGAGUUUGCUCAGCGGCUGUUUCGACAUGAGCU ..------.(((((((.......(((((...(.((((((.......--..)))))).).......((....)))))))(((((.((((....)))).)))))..........))))))) ( -31.80) >DroSim_CAF1 64219 111 + 1 CC------CAGCUCAUUAAAUCAUCGUAAAAGUGUCCGCUACCGAG--UUGCGGACUCAAUAUUCUGCUCACAUACGACCGUUUAGCGAGUUUGCUCAGCGGCUGUUUCGACAUGAGCU ..------.(((((((.......(((((...(.((((((.......--..)))))).).......((....)))))))(((((.((((....)))).)))))..........))))))) ( -31.80) >DroEre_CAF1 70114 111 + 1 CC------CAGCUCAUUAAAUCAUCGUAAAAUUGUCCGCUACCGAG--UUGCGGACUCAAUAUUCUCCUCACAUACGACCGUUUAGCGAGUUUGCUCAGCGGCUGUUUCGACAUGAGCU ..------.(((((((.................((((((.......--..))))))..................(((.(((((.((((....)))).))))).)))......))))))) ( -29.80) >DroYak_CAF1 69730 117 + 1 CCCGGUGCCAGCUCAUUAAAUCAUCGUAAAAGUGUCCGCUACCGAG--UUGCGGACUCAAUAUUCUGCUCACAUACGACCGUUUAGCGAGUUUGCUCAGCGGCUGUUUCGCCAUGAGCU .........(((((.((((((..(((((...(.((((((.......--..)))))).).......((....)))))))..)))))).))))).(((((..(((......))).))))). ( -35.10) >DroAna_CAF1 67641 113 + 1 GC------CGGAUCAUUAAAUCAUCGUAAAAGUGUCCGCUACCGACUUUUGCGGACUCAAUAUUCAGCUCACAUACAACCGUUUAGCGAGUUUGCUCAGCUGCUAUUUCAACAUGAGCU ..------.......................(.((((((...........)))))).).......((((((............(((((((....))).))))...........)))))) ( -24.10) >consensus CC______CAGCUCAUUAAAUCAUCGUAAAAGUGUCCGCUACCGAG__UUGCGGACUCAAUAUUCUGCUCACAUACGACCGUUUAGCGAGUUUGCUCAGCGGCUGUUUCGACAUGAGCU .........(((((((...............(.((((((...........)))))).)................(((.(((((.((((....)))).))))).)))......))))))) (-27.25 = -27.50 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:34 2006