
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,619,946 – 4,620,079 |
| Length | 133 |
| Max. P | 0.988589 |

| Location | 4,619,946 – 4,620,065 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.04 |
| Mean single sequence MFE | -35.27 |
| Consensus MFE | -27.62 |
| Energy contribution | -28.28 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
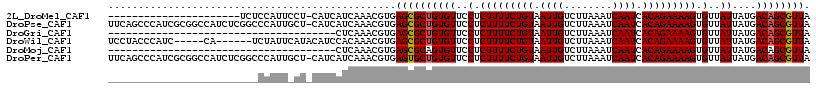
>2L_DroMel_CAF1 4619946 119 + 22407834 UGGUCCUGGG-CAAUUUGCAGAGUGCGCUGUCACGAUUAAUAACGCUGUCAUAAUAACACUUUUCUGUGAUUGAUUUAAGACAAUUACAGAAAAGAGGAACACAGCGCUCACGUUUGAUG ..(((....(-(.....)).((((((..(((..((((..........))).........((((((((((((((........)))))))))))))).)..)))..))))))......))). ( -33.00) >DroVir_CAF1 61565 117 + 1 U-GUUCGGCG-CAAUUUGCGCUUUGCGCUGUCACGAUUAAUAACGCUGUCAUAAUAACACUUUUCUGUGAUUGAUUUAAGACAAUUACAGAAAAGAGGAACACAGCGCUCACGUUUGAG- .-((.(((((-(((........))))))))..)).........((((((........(.((((((((((((((........)))))))))))))).)....))))))((((....))))- ( -37.90) >DroGri_CAF1 60181 118 + 1 U-GUUUGGCGGUAAUUUGCGCUGUGCGCUGUCACGAUUAAUAACGCUGUCAUAAUAACACUUUUCUGUGAUUGAUUUAAGACAAUUACAGAAAAGAGGAACACAGCGCUCACGUUUGAG- .-.((..(((.......((((((((..(((.((((........)).)).).........((((((((((((((........)))))))))))))).))..))))))))...)))..)).- ( -36.80) >DroWil_CAF1 54229 117 + 1 UUGUCCAUGG-CAAUUUGCGC--CGCGCUGUCACGAUUAAUAACGCUGUCAUAAUAACACUUUUCUGUGAUUGAUUUAAGACAAUUACAGAAAAGAGGAACACAGCGCUCACGUUUGUGG ....((((((-(.......))--)(((((((...(((..........))).........((((((((((((((........))))))))))))))......)))))))........)))) ( -36.30) >DroMoj_CAF1 70847 117 + 1 U-GUUCGGCG-CAAUUUGCGCUUUGCGCUGUCACGAUUAAUAACGCUGUCAUAAUAACACUUUUCUGUGAUUGAUUUAAGACAAUUACAGAAAAGAGGAACACUGCGCUCACGUUUGAG- (-((((((((-(((........)))))))(.((((........)).)).).........((((((((((((((........))))))))))))))..))))).....((((....))))- ( -33.90) >DroPer_CAF1 63476 119 + 1 CAGUCUGGGG-CAAUUUGCAGAGUGCGCUGUCACGAUUAAUAACGCUGUCAUAAUAACACUUUUCUGUGAUUGAUUUAAGACAAUUACAGAAAAGAGGAACACAGCACUCACGUUUGAUG ..(((....(-(.....)).((((((..(((..((((..........))).........((((((((((((((........)))))))))))))).)..)))..))))))......))). ( -33.70) >consensus U_GUCCGGCG_CAAUUUGCGCUGUGCGCUGUCACGAUUAAUAACGCUGUCAUAAUAACACUUUUCUGUGAUUGAUUUAAGACAAUUACAGAAAAGAGGAACACAGCGCUCACGUUUGAG_ .................(((....(((((((...(((..........))).........((((((((((((((........))))))))))))))......)))))))...)))...... (-27.62 = -28.28 + 0.67)


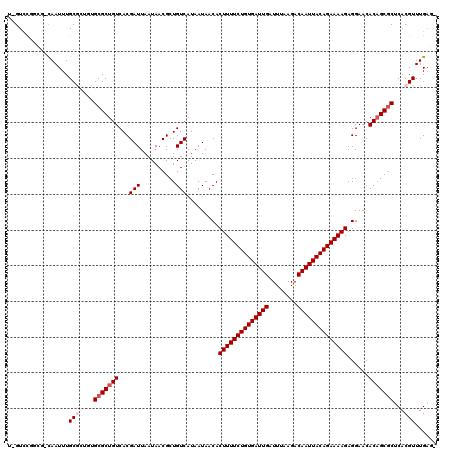
| Location | 4,619,946 – 4,620,065 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.04 |
| Mean single sequence MFE | -32.87 |
| Consensus MFE | -28.33 |
| Energy contribution | -28.13 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4619946 119 - 22407834 CAUCAAACGUGAGCGCUGUGUUCCUCUUUUCUGUAAUUGUCUUAAAUCAAUCACAGAAAAGUGUUAUUAUGACAGCGUUAUUAAUCGUGACAGCGCACUCUGCAAAUUG-CCCAGGACCA ........(((.(((((((((..(.(((((((((.((((........)))).))))))))).)..))....)))))(((((.....))))).)).)))(((((.....)-)...)))... ( -30.30) >DroVir_CAF1 61565 117 - 1 -CUCAAACGUGAGCGCUGUGUUCCUCUUUUCUGUAAUUGUCUUAAAUCAAUCACAGAAAAGUGUUAUUAUGACAGCGUUAUUAAUCGUGACAGCGCAAAGCGCAAAUUG-CGCCGAAC-A -......((...(((((((((..(.(((((((((.((((........)))).))))))))).)..)).......(((........))).)))))))...((((.....)-)))))...-. ( -34.90) >DroGri_CAF1 60181 118 - 1 -CUCAAACGUGAGCGCUGUGUUCCUCUUUUCUGUAAUUGUCUUAAAUCAAUCACAGAAAAGUGUUAUUAUGACAGCGUUAUUAAUCGUGACAGCGCACAGCGCAAAUUACCGCCAAAC-A -...........(((((((((....(((((((((.((((........)))).)))))))))((((.....))))(((((((.....))))).)))))))))))...............-. ( -34.90) >DroWil_CAF1 54229 117 - 1 CCACAAACGUGAGCGCUGUGUUCCUCUUUUCUGUAAUUGUCUUAAAUCAAUCACAGAAAAGUGUUAUUAUGACAGCGUUAUUAAUCGUGACAGCGCG--GCGCAAAUUG-CCAUGGACAA (((.........(((((((((..(.(((((((((.((((........)))).))))))))).)..)).......(((........))).)))))))(--(((.....))-)).))).... ( -35.10) >DroMoj_CAF1 70847 117 - 1 -CUCAAACGUGAGCGCAGUGUUCCUCUUUUCUGUAAUUGUCUUAAAUCAAUCACAGAAAAGUGUUAUUAUGACAGCGUUAUUAAUCGUGACAGCGCAAAGCGCAAAUUG-CGCCGAAC-A -.......((..(((((((......(((((((((.((((........)))).)))))))))((((((.(((((...))))).....))))))((((...))))..))))-)))...))-. ( -32.80) >DroPer_CAF1 63476 119 - 1 CAUCAAACGUGAGUGCUGUGUUCCUCUUUUCUGUAAUUGUCUUAAAUCAAUCACAGAAAAGUGUUAUUAUGACAGCGUUAUUAAUCGUGACAGCGCACUCUGCAAAUUG-CCCCAGACUG ........(((((((((((((..(.(((((((((.((((........)))).))))))))).)..))....)))(((((((.....))))).)))))))).))......-.......... ( -29.20) >consensus _CUCAAACGUGAGCGCUGUGUUCCUCUUUUCUGUAAUUGUCUUAAAUCAAUCACAGAAAAGUGUUAUUAUGACAGCGUUAUUAAUCGUGACAGCGCACAGCGCAAAUUG_CCCCGAAC_A ......(((..((((((((((..(.(((((((((.((((........)))).))))))))).)..))....))))))))......)))....(((.....)))................. (-28.33 = -28.13 + -0.19)

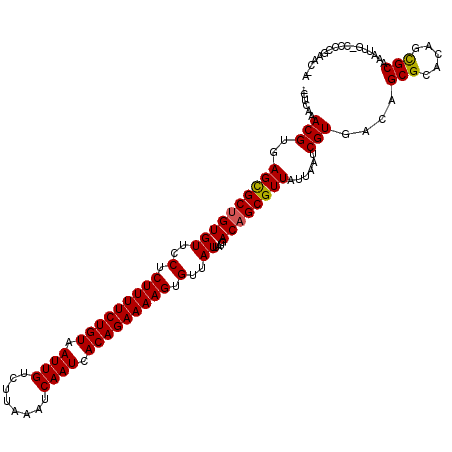
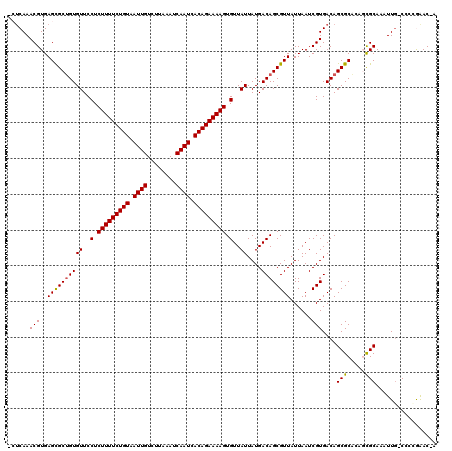
| Location | 4,619,985 – 4,620,079 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -22.23 |
| Energy contribution | -22.57 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
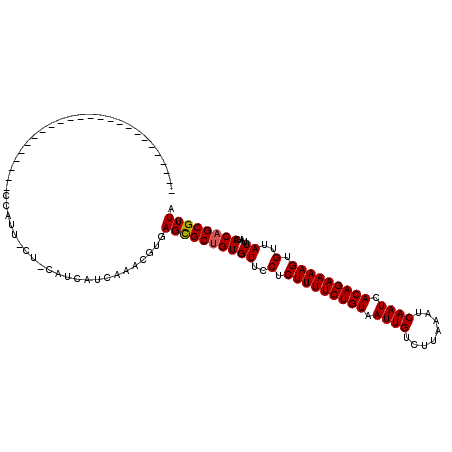
>2L_DroMel_CAF1 4619985 94 + 22407834 UAACGCUGUCAUAAUAACACUUUUCUGUGAUUGAUUUAAGACAAUUACAGAAAAGAGGAACACAGCGCUCACGUUUGAUGAUG-AGGAAUGGAGA---------------------- ...((((((........(.((((((((((((((........)))))))))))))).)....))))))(((.(((((.......-..)))))))).---------------------- ( -28.10) >DroPse_CAF1 71845 116 + 1 UAACGCUGUCAUAAUAACACUUUUCUGUGAUUGAUUUAAGACAAUUACAGAAAAGAGGAACACAGCGCUCACGUUUGAUGAUG-AGCAAUGGGCCGAGAUGGCCGCGAUGGGCUGAA .....(((((.........((((((((((((((........)))))))))))))).........((((((((((...))).))-)))....((((.....)))))))))))...... ( -38.50) >DroGri_CAF1 60220 79 + 1 UAACGCUGUCAUAAUAACACUUUUCUGUGAUUGAUUUAAGACAAUUACAGAAAAGAGGAACACAGCGCUCACGUUUGAG-------------------------------------- ...((((((........(.((((((((((((((........)))))))))))))).)....))))))((((....))))-------------------------------------- ( -25.30) >DroWil_CAF1 54266 106 + 1 UAACGCUGUCAUAAUAACACUUUUCUGUGAUUGAUUUAAGACAAUUACAGAAAAGAGGAACACAGCGCUCACGUUUGUGGAUGUAUGAAUAGA------UG-----GAUGGGUAGGA ....(((((........(.((((((((((((((........)))))))))))))).)....)))))((((((((((((..........)))))------))-----..))))).... ( -30.00) >DroMoj_CAF1 70885 79 + 1 UAACGCUGUCAUAAUAACACUUUUCUGUGAUUGAUUUAAGACAAUUACAGAAAAGAGGAACACUGCGCUCACGUUUGAG-------------------------------------- ...(((.............((((((((((((((........))))))))))))))((.....)))))((((....))))-------------------------------------- ( -21.30) >DroPer_CAF1 63515 116 + 1 UAACGCUGUCAUAAUAACACUUUUCUGUGAUUGAUUUAAGACAAUUACAGAAAAGAGGAACACAGCACUCACGUUUGAUGAUG-AGCAAUGGGCCGAGAUGGCCGCGAUGGGCUGAA ...(((.(((((.......((((((((((((((........)))))))))))))).((..((..((.((((.......))).)-.))..))..))...))))).))).......... ( -34.70) >consensus UAACGCUGUCAUAAUAACACUUUUCUGUGAUUGAUUUAAGACAAUUACAGAAAAGAGGAACACAGCGCUCACGUUUGAGGAUG_AG_AAUGG_________________________ ...((((((........(.((((((((((((((........)))))))))))))).)....)))))).................................................. (-22.23 = -22.57 + 0.33)

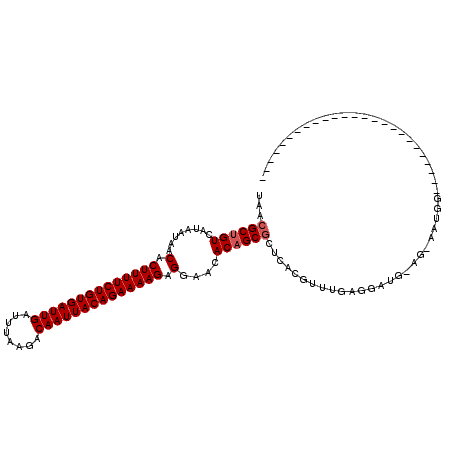
| Location | 4,619,985 – 4,620,079 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -23.37 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.83 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4619985 94 - 22407834 ----------------------UCUCCAUUCCU-CAUCAUCAAACGUGAGCGCUGUGUUCCUCUUUUCUGUAAUUGUCUUAAAUCAAUCACAGAAAAGUGUUAUUAUGACAGCGUUA ----------------------.........((-(((........)))))((((((((..(.(((((((((.((((........)))).))))))))).)..))....))))))... ( -25.20) >DroPse_CAF1 71845 116 - 1 UUCAGCCCAUCGCGGCCAUCUCGGCCCAUUGCU-CAUCAUCAAACGUGAGCGCUGUGUUCCUCUUUUCUGUAAUUGUCUUAAAUCAAUCACAGAAAAGUGUUAUUAUGACAGCGUUA ....((.....))((((.....))))....(((-(((........))))))(((((((..(.(((((((((.((((........)))).))))))))).)..))....))))).... ( -36.00) >DroGri_CAF1 60220 79 - 1 --------------------------------------CUCAAACGUGAGCGCUGUGUUCCUCUUUUCUGUAAUUGUCUUAAAUCAAUCACAGAAAAGUGUUAUUAUGACAGCGUUA --------------------------------------..........((((((((((..(.(((((((((.((((........)))).))))))))).)..))....)))))))). ( -24.90) >DroWil_CAF1 54266 106 - 1 UCCUACCCAUC-----CA------UCUAUUCAUACAUCCACAAACGUGAGCGCUGUGUUCCUCUUUUCUGUAAUUGUCUUAAAUCAAUCACAGAAAAGUGUUAUUAUGACAGCGUUA ...........-----..------..............(((....)))((((((((((..(.(((((((((.((((........)))).))))))))).)..))....)))))))). ( -26.50) >DroMoj_CAF1 70885 79 - 1 --------------------------------------CUCAAACGUGAGCGCAGUGUUCCUCUUUUCUGUAAUUGUCUUAAAUCAAUCACAGAAAAGUGUUAUUAUGACAGCGUUA --------------------------------------..........(((((.........(((((((((.((((........)))).)))))))))((((.....))))))))). ( -20.40) >DroPer_CAF1 63515 116 - 1 UUCAGCCCAUCGCGGCCAUCUCGGCCCAUUGCU-CAUCAUCAAACGUGAGUGCUGUGUUCCUCUUUUCUGUAAUUGUCUUAAAUCAAUCACAGAAAAGUGUUAUUAUGACAGCGUUA ....((.....))((((.....))))....(((-(((........))))))(((((((..(.(((((((((.((((........)))).))))))))).)..))....))))).... ( -33.50) >consensus _________________________CCAUU_CU_CAUCAUCAAACGUGAGCGCUGUGUUCCUCUUUUCUGUAAUUGUCUUAAAUCAAUCACAGAAAAGUGUUAUUAUGACAGCGUUA ................................................((((((((((..(.(((((((((.((((........)))).))))))))).)..))....)))))))). (-23.37 = -23.40 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:32 2006