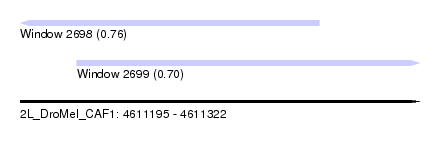
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,611,195 – 4,611,322 |
| Length | 127 |
| Max. P | 0.756651 |

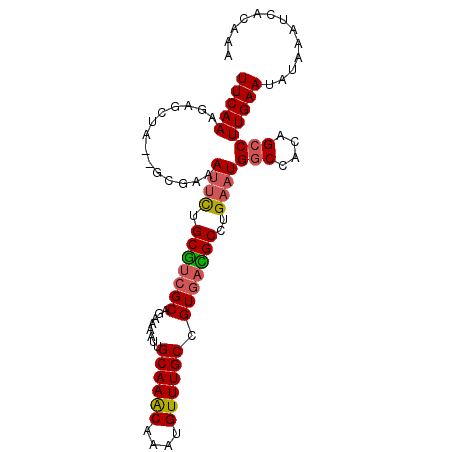
| Location | 4,611,195 – 4,611,290 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 88.22 |
| Mean single sequence MFE | -22.65 |
| Consensus MFE | -19.25 |
| Energy contribution | -19.45 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4611195 95 - 22407834 UUCAAAGAGCUA--GCGAAAUUCUGCGUCGCAGAAAAUUGCAAACAAAUGUUUGCCGUGACGCCUGAAUGGCCACAGCCUUGAAUAUAAAUCACAAA (((((.(.(((.--((...((((.(((((((........((((((....)))))).)))))))..)))).))...)))))))))............. ( -25.10) >DroSec_CAF1 48585 95 - 1 UUCAAAGAGCUA--GCGAAAUUCUGCGUCGCAGAAAAUUGCAAACAAAUGUUUGCCGUGACGCCUGAAUGGCCACAGCCUUGAAUAUAAAUCACAAA (((((.(.(((.--((...((((.(((((((........((((((....)))))).)))))))..)))).))...)))))))))............. ( -25.10) >DroSim_CAF1 46230 95 - 1 UUCAAAGAGCUA--GCGAAAUUCUGCGUCGCAGAAAAUUGCAAACAAAUGUUUGCCGUGACGCCUGAAUGGCCACAGCCUUGAAUAUAAAUCACAAA (((((.(.(((.--((...((((.(((((((........((((((....)))))).)))))))..)))).))...)))))))))............. ( -25.10) >DroEre_CAF1 51210 95 - 1 UUCAAAGAGCUA--ACGAAAUUCUGCGUCGCAAAAAAUUGCAAACAAAUGUUUGCCGUGACGCCUGAAUGGCCACAGCCUUGAAUUUAAAUCACAAA (((((.(.(((.--..(..((((.(((((((........((((((....)))))).)))))))..))))..)...)))))))))............. ( -25.00) >DroAna_CAF1 49180 95 - 1 UUCAAAGGUUUA--UCGAAAUUUUGCAACGCAGGGAGUUGCAAACAAAUGUUUGCCGUGAUGCCUGAAUGACUACAGCCUUGAAUAUAAAUCAGAUG (((((.((((..--.....((((.(((.(((........((((((....)))))).))).)))..))))......)))))))))............. ( -17.32) >DroPer_CAF1 54018 97 - 1 UUCAAAAGAGUAAUUCGCAAAACAGCGUCGCAGAGAAUUGCAAGCAAAUGUUUGCCGUAAUGCCUGAAUGGCCACAGCCUUGAAUUUAAAUCAGUAA (((((.......((((((......))...(((....((.((((((....)))))).))..)))..))))(((....))))))))............. ( -18.30) >consensus UUCAAAGAGCUA__GCGAAAUUCUGCGUCGCAGAAAAUUGCAAACAAAUGUUUGCCGUGACGCCUGAAUGGCCACAGCCUUGAAUAUAAAUCACAAA (((((..............((((.(((((((........((((((....)))))).)))))))..))))(((....))))))))............. (-19.25 = -19.45 + 0.20)

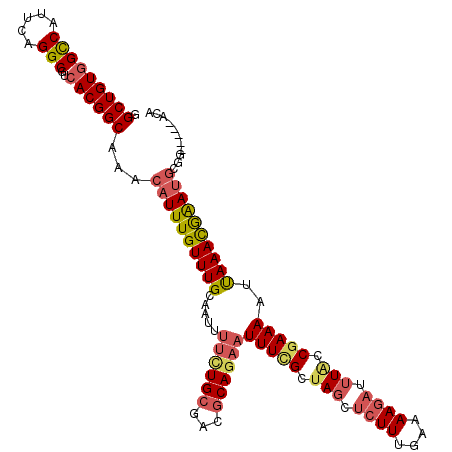
| Location | 4,611,213 – 4,611,322 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 88.98 |
| Mean single sequence MFE | -30.95 |
| Consensus MFE | -25.72 |
| Energy contribution | -26.70 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4611213 109 + 22407834 GGCUGUGGCCAUUCAGGCGUCACGGCAAACAUUUGUUUGCAAUUUUCUGCGACGCAGAAUUUCGCUAGCUCUUUGAAAAGAUUUGCCGAAAAUUAAACAAAUGCGG-----ACA .(((((((((.....)))..)))))).......((((((((...((((((...))))))(((((.(((.((((....)))).))).)))))..........)))))-----))) ( -33.00) >DroSec_CAF1 48603 109 + 1 GGCUGUGGCCAUUCAGGCGUCACGGCAAACAUUUGUUUGCAAUUUUCUGCGACGCAGAAUUUCGCUAGCUCUUUGAAAAGAUUUACCGAAAAUUAAACGAAUGCGG-----ACA .(((((((((.....)))..)))))).......((((((((...((((((...))))))(((((.(((.((((....)))).))).)))))..........)))))-----))) ( -33.30) >DroSim_CAF1 46248 109 + 1 GGCUGUGGCCAUUCAGGCGUCACGGCAAACAUUUGUUUGCAAUUUUCUGCGACGCAGAAUUUCGCUAGCUCUUUGAAAAGAUUUACCGAAAAUUAAACGAAUGCGG-----ACA .(((((((((.....)))..)))))).......((((((((...((((((...))))))(((((.(((.((((....)))).))).)))))..........)))))-----))) ( -33.30) >DroEre_CAF1 51228 109 + 1 GGCUGUGGCCAUUCAGGCGUCACGGCAAACAUUUGUUUGCAAUUUUUUGCGACGCAGAAUUUCGUUAGCUCUUUGAAAAGAUUUACCGAAAAUUAAACGAAUGCGG-----ACA .(((((((((.....)))..)))))).......((((((((...((((((...))))))(((((.(((.((((....)))).))).)))))..........)))))-----))) ( -30.70) >DroYak_CAF1 51833 109 + 1 GGCUGUGGCCAUUCAGGCGUCACGGCAAACAUUUGUUUGCCAUUUUCUGAGACGCAGAAUUUUGCUAGCUCUUUGAAAAGAUUUACCGAAAAUUAAACGAAUGCGG-----ACA .(((((((((.....)))..))))))...((((((((((..((((((.(((..((((....))))...)))(((....)))......))))))))))))))))...-----... ( -29.40) >DroAna_CAF1 49198 114 + 1 GGCUGUAGUCAUUCAGGCAUCACGGCAAACAUUUGUUUGCAACUCCCUGCGUUGCAAAAUUUCGAUAAACCUUUGAAAAAAUCUCCCAAAAACCAAAUGGAAACGUGUUCCGCA (((....))).....((...((((.....((((((((((((((.......)))))))).((((((.......))))))...............))))))....))))..))... ( -26.00) >consensus GGCUGUGGCCAUUCAGGCGUCACGGCAAACAUUUGUUUGCAAUUUUCUGCGACGCAGAAUUUCGCUAGCUCUUUGAAAAGAUUUACCGAAAAUUAAACGAAUGCGG_____ACA .(((((((((.....)))..))))))...((((((((((.....((((((...))))))(((((.(((.((((....)))).))).)))))..))))))))))........... (-25.72 = -26.70 + 0.98)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:28 2006