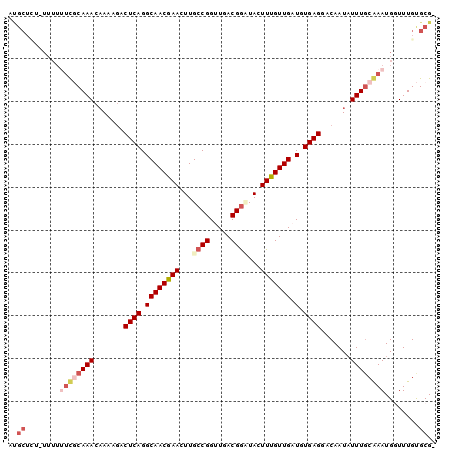
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,573,139 – 4,573,235 |
| Length | 96 |
| Max. P | 0.680386 |

| Location | 4,573,139 – 4,573,235 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 84.65 |
| Mean single sequence MFE | -21.48 |
| Consensus MFE | -15.58 |
| Energy contribution | -16.18 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4573139 96 + 22407834 -CGCACAAAACAUUUGCAAAUAUUGUCCUCACAUCAACAAAAUAGCCGUCAACCGGCAAGUUCGUUGCCUGAGUCUUUUGUUUGCA--AAAA-ACAGCAU -...........((((((((((..(..((((...((((.((...((((.....))))...)).))))..))))..)..))))))))--))..-....... ( -24.30) >DroSec_CAF1 17965 98 + 1 -CGCACAAACCAUUCGCAAAUAUUGUCCUCACAUCAACAAAGUAGCCGUCAACCGGCAAGUUCGUUGCCUGAGUCUUUGGUUUGCGAAAAAA-AGAGCAU -.((........(((((((((...(..((((...((((.((.(.((((.....)))).).)).))))..))))..)...)))))))))....-...)).. ( -25.86) >DroSim_CAF1 15091 98 + 1 -CGCACAAAACAUUCGCAAAUAUUGUCCUCACAUCAACAAAGUAACCGUCAACCGGCAAGUUCGUUGCCUGAGUCUUUUGUUUGCGAAAAAA-AGAGCGU -(((........((((((((((..(..((((...((((.((.(..(((.....)))..).)).))))..))))..)..))))))))))....-...))). ( -24.46) >DroEre_CAF1 20467 87 + 1 CCGCGCAAACCAUUUGUAAAAAUUGUCCUCAGAACAACAAAGUAUUCGUCAACCGGCAAGUUUGUUGCCUGAGUUUUUUGUUUGAAA------------- .....(((((.((((....))))....(((((..(((((((.(..(((.....)))..).))))))).)))))......)))))...------------- ( -15.20) >DroYak_CAF1 18117 100 + 1 UCGCGCAAACCAUUUGCAAAUAUUGUCCUCAAAACAACAAAGUAUCCGUCAACCGGCAAGUUCGUUGCCUGAGUCUUUUGUUUGAGAAAAAAUAAAGCAU ..((((((.....))))...((((...(((((....((((((.((...(((...(((((.....)))))))))).)))))))))))....))))..)).. ( -17.60) >consensus _CGCACAAACCAUUUGCAAAUAUUGUCCUCACAUCAACAAAGUAGCCGUCAACCGGCAAGUUCGUUGCCUGAGUCUUUUGUUUGCGAAAAAA_AGAGCAU ............((((((((((..(..((((...((((.((.(..(((.....)))..).)).))))..))))..)..))))))))))............ (-15.58 = -16.18 + 0.60)


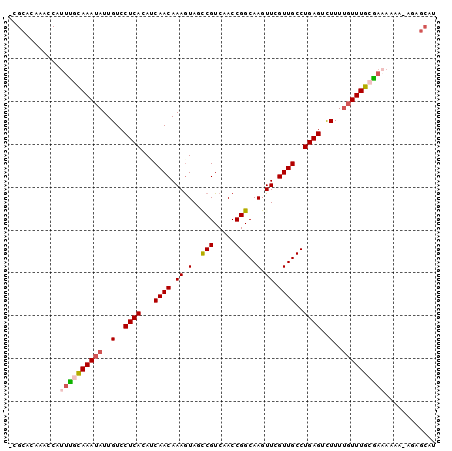
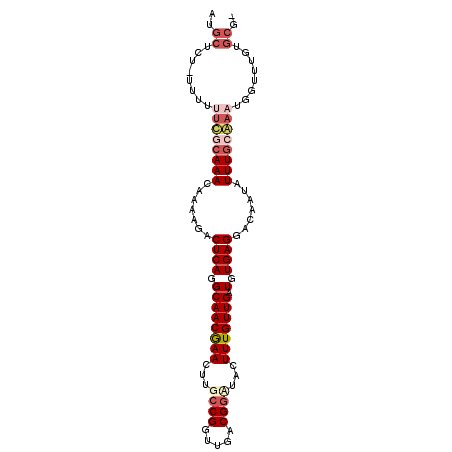
| Location | 4,573,139 – 4,573,235 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 84.65 |
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -20.25 |
| Energy contribution | -22.09 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.680386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4573139 96 - 22407834 AUGCUGU-UUUU--UGCAAACAAAAGACUCAGGCAACGAACUUGCCGGUUGACGGCUAUUUUGUUGAUGUGAGGACAAUAUUUGCAAAUGUUUUGUGCG- ..((...-..((--((((((.......((((.((((((((...((((.....))))...))))))).).)))).......))))))))........)).- ( -28.66) >DroSec_CAF1 17965 98 - 1 AUGCUCU-UUUUUUCGCAAACCAAAGACUCAGGCAACGAACUUGCCGGUUGACGGCUACUUUGUUGAUGUGAGGACAAUAUUUGCGAAUGGUUUGUGCG- ..((.(.-.(..((((((((.......((((.((((((((...((((.....))))...))))))).).)))).......))))))))..)...).)).- ( -32.04) >DroSim_CAF1 15091 98 - 1 ACGCUCU-UUUUUUCGCAAACAAAAGACUCAGGCAACGAACUUGCCGGUUGACGGUUACUUUGUUGAUGUGAGGACAAUAUUUGCGAAUGUUUUGUGCG- .(((.(.-....((((((((.......((((.((((((((...((((.....))))...))))))).).)))).......))))))))......).)))- ( -31.54) >DroEre_CAF1 20467 87 - 1 -------------UUUCAAACAAAAAACUCAGGCAACAAACUUGCCGGUUGACGAAUACUUUGUUGUUCUGAGGACAAUUUUUACAAAUGGUUUGCGCGG -------------...(((((......(((((((((((((.....((.....)).....))))))).))))))...........(....))))))..... ( -17.50) >DroYak_CAF1 18117 100 - 1 AUGCUUUAUUUUUUCUCAAACAAAAGACUCAGGCAACGAACUUGCCGGUUGACGGAUACUUUGUUGUUUUGAGGACAAUAUUUGCAAAUGGUUUGCGCGA .(((.......................(((((((((((((....(((.....)))....))))))).))))))..........((((.....))))))). ( -22.70) >consensus AUGCUCU_UUUUUUCGCAAACAAAAGACUCAGGCAACGAACUUGCCGGUUGACGGAUACUUUGUUGAUGUGAGGACAAUAUUUGCAAAUGGUUUGUGCG_ ..((........((((((((.......((((.((((((((...((((.....))))...))))))).).)))).......))))))))........)).. (-20.25 = -22.09 + 1.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:18 2006