| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,532,087 – 4,532,220 |
| Length | 133 |
| Max. P | 0.639736 |

| Location | 4,532,087 – 4,532,198 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.81 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -20.76 |
| Energy contribution | -20.65 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
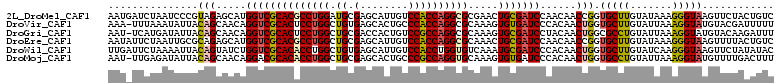
>2L_DroMel_CAF1 4532087 111 - 22407834 AAUGAUCUAAUCCCGUAGAGCAUGGUCGCACGCCUGGAUGCGAGCAUUGUCCACCAGGCGCGAACUGCGAUCCAACAACCGGUGCUUGUAUAAAGGGUAAGUUCUACUGUC ..............(((((((.(((((((((((((((.((.((......))))))))))).....))))).)))....((.(((....)))...))....))))))).... ( -36.10) >DroVir_CAF1 48088 110 - 1 AAA-UUUAAAUAUUACAGCAACAGGUCGCACUCCUGGCUGUGAGCACUGCCCACCAGGCGCAAAGUGUGAUCCCACAACUGGUGCUUGUAUUAAAGGUAUGUACGAUUUUU ...-........(((((((..((((.......)))))))))))(((.((((...(((((((....((((....))))....))))))).......)))))))......... ( -31.30) >DroGri_CAF1 52084 110 - 1 AAU-UCAUGAUAUUACAGCAACAGGUCGCACUCCUGGCUGCGACCACUGUCCGCCAGGCGCAAAGUGCGAUCCUACAACUGGCGCCUGUAUUAAAGGUAUGUACAAGAUUU ...-((.((((((..(((....((((((((((((((((.(.(((....)))))))))).....))))))).)))....)))..((((.......)))))))).)).))... ( -33.90) >DroEre_CAF1 47849 111 - 1 AAUAUUCUAAUUGCGCAGAGCAUGGUCGCACGCCUGGCUGCGAGCAUUGUCCACCAGGCGCAAACUGCGAUCCAACAACCGGUGCUUGUAUAAAGGGUAAGUUUUACUGUC ..((((((..(((..((.(((((((((((((((((((.((.((......))))))))))).....))))))).........)))))))..)))))))))............ ( -32.20) >DroWil_CAF1 2343 111 - 1 UUGAUUCUAAAAUUACAGUAUCUGGUCGCACACCUGGCUGUGAGCAUUGUCCACCUGGUGUCAAAUGCGAUCCCACAACUGGUGCUUGUAUCAAGGGUAAGUUCUAUAUAC ...(((((.....(((((((((.(((((((((((.((.((.((......)))))).))))).....))))))........))))).))))...)))))............. ( -24.20) >DroMoj_CAF1 54931 110 - 1 AAU-UUGAGAUAUUACAGCAACAGGACGCACACCUGGCUGCGAGCACUGCCCGCCAGGUGCAAAGUGUGAUCCCACAACUGGUGCCUGUAUUAAAGGUAUGUUUUGACUUU ...-(..((((((.((....(((((.(((.((((((((.(((.....)))..))))))))...((((((....))).))).)))))))).......))))))))..).... ( -35.00) >consensus AAU_UUCUAAUAUUACAGCAACAGGUCGCACACCUGGCUGCGAGCACUGUCCACCAGGCGCAAAGUGCGAUCCCACAACUGGUGCUUGUAUUAAAGGUAAGUUCUACUUUC ...............(((.....((((((((((((((.((.(........)))))))))).....)))))))......))).(((((.......)))))............ (-20.76 = -20.65 + -0.11)
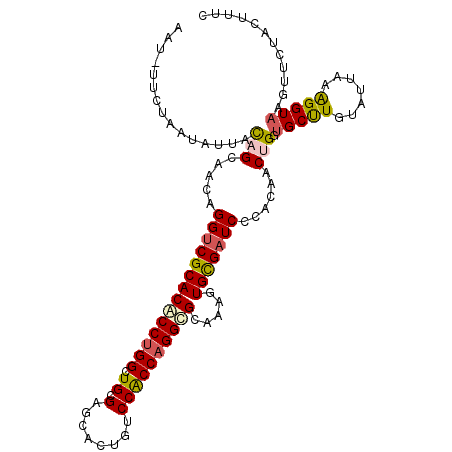
| Location | 4,532,125 – 4,532,220 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 83.16 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -22.88 |
| Energy contribution | -23.04 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4532125 95 - 22407834 UGAAUAACUGGCUCAAACUAUUAAUGAUCUAAUCCCGUAGAGCAUGGUCGCACGCCUGGAUGCGAGCAUUGUCCACCAGGCGCGAACUGCGAUCC ..........((((.....((((......))))......))))..((((((((((((((.((.((......))))))))))).....))))))). ( -27.50) >DroVir_CAF1 48126 93 - 1 -AAAUUAUUGGAUUGUGUUAUUAAA-UUUAAAUAUUACAGCAACAGGUCGCACUCCUGGCUGUGAGCACUGCCCACCAGGCGCAAAGUGUGAUCC -........(((((...........-........(((((((..((((.......)))))))))))(((((((((....)).))..)))))))))) ( -25.10) >DroSec_CAF1 47137 95 - 1 UUAAUAACUGGCUCAAAUUAUAAAUCUUCUAAUCUCGCAGAGCAUGGUCGCACGCCUGGAUGCGGGCAUUGCCCACCAGGCGCGAACUGCGAUCC ..........((((.........................))))..((((((((((((((....((((...)))).))))))).....))))))). ( -30.71) >DroSim_CAF1 12017 95 - 1 UGAAUAACCGGCUCAAAUUAUGAAUGUUCUAAUCCCGCAGAGCAUGGUCGCACGCCUGGAUGCGAGCAUUGUCCACCAGGCGCGAACUGCGAUCC .......................(((((((........)))))))((((((((((((((.((.((......))))))))))).....))))))). ( -29.30) >DroEre_CAF1 47887 95 - 1 UAAAUAACUGGUUCAAUUUAUUAAUAUUCUAAUUGCGCAGAGCAUGGUCGCACGCCUGGCUGCGAGCAUUGUCCACCAGGCGCAAACUGCGAUCC .......(((...(((((............)))))..))).....((((((((((((((.((.((......))))))))))).....))))))). ( -25.90) >DroYak_CAF1 39066 95 - 1 UGAAUAACUGGUUCAAUUUAUUAAUAUUCUAAUCCCGCAGAGCACGGUCGCACGCCUGGCUGCGAACAUUGUCCACCAGGCGCAAACUGCGAUCC (((((.....)))))....................(((((.((......)).(((((((.((.((......)))))))))))....))))).... ( -26.70) >consensus UGAAUAACUGGCUCAAAUUAUUAAUAUUCUAAUCCCGCAGAGCAUGGUCGCACGCCUGGAUGCGAGCAUUGUCCACCAGGCGCAAACUGCGAUCC ..........((((.........................))))..((((((((((((((.((.((......))))))))))).....))))))). (-22.88 = -23.04 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:09 2006