
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,525,243 – 4,525,357 |
| Length | 114 |
| Max. P | 0.596962 |

| Location | 4,525,243 – 4,525,357 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.59 |
| Mean single sequence MFE | -44.42 |
| Consensus MFE | -24.21 |
| Energy contribution | -25.41 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
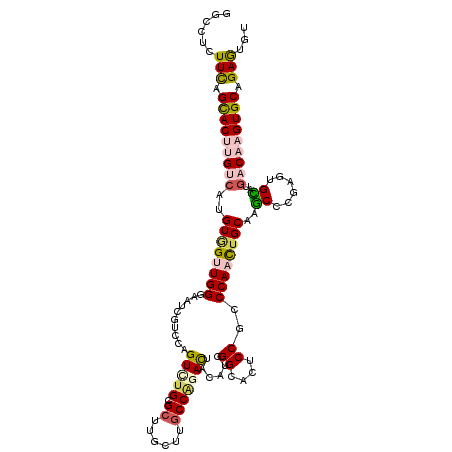
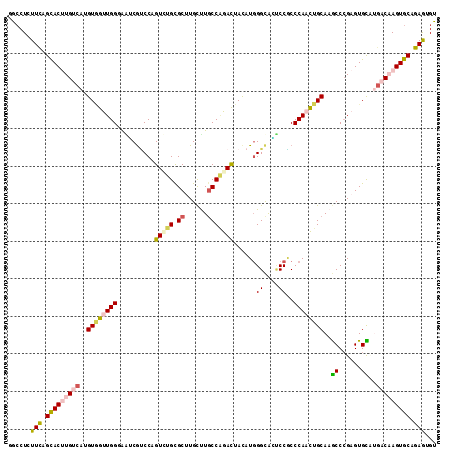
>2L_DroMel_CAF1 4525243 114 + 22407834 ACACUCGGCACUGGUCAUACACUCGGGCUUGCAGUUGGGCGGAGCACCCAUGUAGUCUGGCAAGCAGGCACACACAGGACGCUCCCCAACUACAUGACAAGUGCUGAACAGGCC ....((((((((.(((((........(....)(((((((..((((..((.(((.(((((.....)))))....)))))..)))))))))))..))))).))))))))....... ( -41.50) >DroVir_CAF1 40596 114 + 1 ACACUCUGCACUUGUCAAACACUCCGGCUUGCAGUUGGGUGGGUUGCCCAUAUAAUCUGGCAGGCAAGCGCAGACUGGACGACUCCCAACAACAUGACAGGUGCUAAAGAGGCC ...((((((((((((((..(((.(((((.....))))))))(((((.(((.....((((((......)).)))).))).)))))..........))))))))))...))))... ( -41.50) >DroPse_CAF1 41202 114 + 1 ACAUUCUGCACUUGUGCGGCACUCGGGCUUGCAGUUGGGCGGAACGCCAAGGUAGUCCGGAAGGCAAGCGCAAACGGGGUGAUCUCCAGCCACGUGGCAAGUGCUAAACAGGCC ..........(((((..((((((.(.((((((..((((((.....((....)).))))))...)))))).)....((((....)))).(((....))).))))))..))))).. ( -42.80) >DroGri_CAF1 41073 114 + 1 GCACUCUGCACUUGUCAGACACUCGGGUUUGCAAUUGGGCGGACUGCCCAUAUAGUCUGGCAAGCAAGCGCAGACUGGACGACUGCCAACCACAUGACAAGUGCUAAAGAGGCC ((.((((((((((((((........((((.(((..(((((.....)))))..(((((((((......)).)))))))......))).))))...))))))))))...)))))). ( -48.50) >DroMoj_CAF1 47736 114 + 1 ACACUCAGCACUUGUCAAGCACUCGGGCUUGCAGUUGGGUGGGCUGCCCAUAUAGUCGGGCAAACAAGCGCAGACUGGACGGCUACCAACCACUUGGCAUGUACUGAAGAGGCC ....((((.((.((.(((((......)))))))((..(((((...(((....((((((.((......)).).)))))...)))......)))))..))..)).))))....... ( -38.20) >DroAna_CAF1 40597 114 + 1 GCAUUCGGCACUGGUCCUGCACUCGGGCUUGCAGUUGGGAGGCGCGCCCAUGUAGUCUGGCAAGCAGGCGCACACGGGUCGCUCCCCAGCUACGUGACAAGUGCUGAACAGCCC ((.(((((((((.((((.(((........)))(((((((..(((.((((.(((.(((((.....)))))..))).)))))))..)))))))..).))).)))))))))..)).. ( -54.00) >consensus ACACUCUGCACUUGUCAAACACUCGGGCUUGCAGUUGGGCGGACCGCCCAUAUAGUCUGGCAAGCAAGCGCAGACUGGACGACCCCCAACCACAUGACAAGUGCUAAACAGGCC .......((((((((((.......(.((((((...(((((.....))))).............)))))).).....((.......)).......)))))))))).......... (-24.21 = -25.41 + 1.20)



| Location | 4,525,243 – 4,525,357 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.59 |
| Mean single sequence MFE | -44.87 |
| Consensus MFE | -26.33 |
| Energy contribution | -27.03 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4525243 114 - 22407834 GGCCUGUUCAGCACUUGUCAUGUAGUUGGGGAGCGUCCUGUGUGUGCCUGCUUGCCAGACUACAUGGGUGCUCCGCCCAACUGCAAGCCCGAGUGUAUGACCAGUGCCGAGUGU ((((((.((((((((((.(.(((((((((((((((.((((((((.(.(((.....))).))))))))))))))..)))))))))).)..))))))).))).))).)))...... ( -54.30) >DroVir_CAF1 40596 114 - 1 GGCCUCUUUAGCACCUGUCAUGUUGUUGGGAGUCGUCCAGUCUGCGCUUGCCUGCCAGAUUAUAUGGGCAACCCACCCAACUGCAAGCCGGAGUGUUUGACAAGUGCAGAGUGU .((((((...((((.((((((((.((((((.((.((((((((((.((......)))))))....))))).))...)))))).))).((....))...))))).)))))))).)) ( -40.90) >DroPse_CAF1 41202 114 - 1 GGCCUGUUUAGCACUUGCCACGUGGCUGGAGAUCACCCCGUUUGCGCUUGCCUUCCGGACUACCUUGGCGUUCCGCCCAACUGCAAGCCCGAGUGCCGCACAAGUGCAGAAUGU .....((((.(((((((....(((((.((........)).((((.((((((.....((....))..((((...)))).....)))))).)))).))))).))))))).)))).. ( -36.20) >DroGri_CAF1 41073 114 - 1 GGCCUCUUUAGCACUUGUCAUGUGGUUGGCAGUCGUCCAGUCUGCGCUUGCUUGCCAGACUAUAUGGGCAGUCCGCCCAAUUGCAAACCCGAGUGUCUGACAAGUGCAGAGUGC .((((((...((((((((((((.((((.(((((.....((((((.((......))))))))...(((((.....)))))))))).))))))......)))))))))))))).)) ( -48.40) >DroMoj_CAF1 47736 114 - 1 GGCCUCUUCAGUACAUGCCAAGUGGUUGGUAGCCGUCCAGUCUGCGCUUGUUUGCCCGACUAUAUGGGCAGCCCACCCAACUGCAAGCCCGAGUGCUUGACAAGUGCUGAGUGU ......((((((((.((.((((((.((((..((.(......).))(((((((((((((......)))))))...........)))))))))).)))))).)).))))))))... ( -39.30) >DroAna_CAF1 40597 114 - 1 GGGCUGUUCAGCACUUGUCACGUAGCUGGGGAGCGACCCGUGUGCGCCUGCUUGCCAGACUACAUGGGCGCGCCUCCCAACUGCAAGCCCGAGUGCAGGACCAGUGCCGAAUGC .(((.((((.(((((((....((((.((((..(((.((((((((.(.(((.....))).)))))))))..)))..)))).)))).....))))))).))))....)))...... ( -50.10) >consensus GGCCUCUUCAGCACUUGUCAUGUGGUUGGGAAUCGUCCAGUCUGCGCUUGCUUGCCAGACUACAUGGGCACUCCGCCCAACUGCAAGCCCGAGUGCAUGACAAGUGCAGAGUGU ......(((.(((((((((..((((((((..........(((((.((......)))))))......((....))..))))))))..((......))..))))))))).)))... (-26.33 = -27.03 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:06 2006