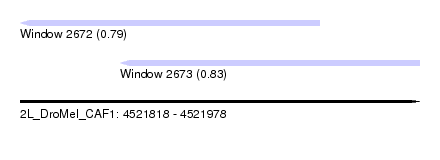
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,521,818 – 4,521,978 |
| Length | 160 |
| Max. P | 0.826431 |

| Location | 4,521,818 – 4,521,938 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -43.40 |
| Consensus MFE | -28.95 |
| Energy contribution | -27.57 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4521818 120 - 22407834 GCUGCCAGGAUCCCUGUCCGGGAUCGUGUGGCUUUAAUGCUUUCUGUAGUGUUGUAAACCACUCGCCCAUCUGCACCUGCGACUCUGGUUACACAGGUGACCCAUUCGCCGGCUGUAACC ((......((((((.....))))))(.((((.(((((((((......))))))).)).)))).))).....((((((.((((....((((((....))))))...)))).)).))))... ( -39.20) >DroPse_CAF1 37844 120 - 1 GCUGCACAGAUCCUUGUCCGGGGUCAUGCGGCUUCAAUGCCUACUGCAGUGUCGUUGGCCACUCGCCUAUCUGCUCGUGCGACAAUGGCUUCACUGGCGAUCCAUUCGCUGGCUGCAAUC ((((((..((((((.....)))))).))))))............(((((.(((((((.((((..((......))..))).).)))))))....(..((((.....))))..))))))... ( -42.80) >DroGri_CAF1 38109 120 - 1 GUUGCAAGGAUCCAUGUCCCGGCUCCUGUGGCUUUAAUGCCUACUGCAACGUAGUGAAUCAUUCACCAGUUUGCACCUGCGAAACCGGCUAUUCUGGCGAUCCAUUUGCUGGCUGCAAUC ((((((((((.((.......)).))))(((((......))).))))))))...((((.....))))....(((((((.(((((..((.(......).)).....))))).)).))))).. ( -35.50) >DroEre_CAF1 37089 120 - 1 GCUGCCAGGAUCCCUGUCCGGGAUCCUGUGGCUUUAAUGCUUACUGCAGUGUGGUAAACCACUCGCCCAUUUGCACCUGCGACACUGGGUACACAGGUGACCCCUUCGCCGGCUGUAACC ((..(.((((((((.....)))))))))..))............((((((((((....))))..(((((.((((....))))...))))).....(((((.....))))).))))))... ( -48.30) >DroAna_CAF1 37357 120 - 1 GCUGCCAGGAUCCUUGUCCGGGAUCCUGUGGCUUCAAUGCCUACUGCAGCGUGGUGAACCACUCGCCCGUCUGCUCUUGCGACAAUGGCUUCACUGGUGAUCCGUUCUCCGGAUGCAAUC ((((((((((((((.....))))))))).(((......)))....)))))((((....))))..(((.(((.((....)))))...)))....((((.((.....)).))))........ ( -49.60) >DroPer_CAF1 39492 120 - 1 GCUGCACAGACCCUUGUCCGGGGUCAUGCGGCUUCAAUGCCUACUGCAGUGUCGUUGGCCACUCGCCCAUCUGUUCGUGCGACAAUGGCUUCACUGGCGAUCCAUUCGCUGGCUGCAAUC ((((((..((((((.....)))))).))))))......(((.....(((((.....(((((.((((.(........).))))...))))).)))))((((.....)))).)))....... ( -45.00) >consensus GCUGCAAGGAUCCUUGUCCGGGAUCCUGUGGCUUCAAUGCCUACUGCAGUGUCGUAAACCACUCGCCCAUCUGCACCUGCGACAAUGGCUACACUGGCGAUCCAUUCGCCGGCUGCAAUC (((((...((((((.....))))))....(((......)))....)))))........((....(((....(((....))).....)))......(((((.....)))))))........ (-28.95 = -27.57 + -1.38)
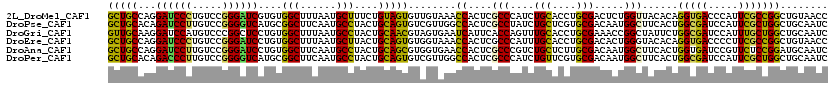

| Location | 4,521,858 – 4,521,978 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.00 |
| Mean single sequence MFE | -43.08 |
| Consensus MFE | -32.46 |
| Energy contribution | -31.30 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4521858 120 - 22407834 AACACAGAGUGUCCUGCCAACCUGGCGUGCAUCAAUGAGCGCUGCCAGGAUCCCUGUCCGGGAUCGUGUGGCUUUAAUGCUUUCUGUAGUGUUGUAAACCACUCGCCCAUCUGCACCUGC ......(((((....((((..(((((((((........)))).)))))((((((.....))))))...))))..(((((((......))))))).....)))))((......))...... ( -41.30) >DroPse_CAF1 37884 120 - 1 AAUUCCGAGUGUCCUGCCAAUCUGGCGUGCAUCAAUGAGCGCUGCACAGAUCCUUGUCCGGGGUCAUGCGGCUUCAAUGCCUACUGCAGUGUCGUUGGCCACUCGCCUAUCUGCUCGUGC .....(((((.....(((.....)))(((..(((((((.(((((((..((((((.....))))))....(((......)))...)))))))))))))).)))..........)))))... ( -42.60) >DroGri_CAF1 38149 120 - 1 AACUCAGAAUGCCCGGCUAAUUUGGCCUGCAUUAACGAACGUUGCAAGGAUCCAUGUCCCGGCUCCUGUGGCUUUAAUGCCUACUGCAACGUAGUGAAUCAUUCACCAGUUUGCACCUGC ....((((((((..((((.....)))).)))))..(((((((((((((((.((.......)).))))(((((......))).))))))))...((((.....))))..)))))...))). ( -37.10) >DroEre_CAF1 37129 120 - 1 AACACCGAGUGUCCUGCAAACCUGGCCUGCAUCAAUGAGCGCUGCCAGGAUCCCUGUCCGGGAUCCUGUGGCUUUAAUGCUUACUGCAGUGUGGUAAACCACUCGCCCAUUUGCACCUGC .((((...))))..((((((...((((((((....(((((((..(.((((((((.....)))))))))..))......))))).))))).((((....))))..)))..))))))..... ( -44.60) >DroAna_CAF1 37397 120 - 1 AACACUGAGUGUCCUGCCACUCUGGCCUGCAUUAAUGAGCGCUGCCAGGAUCCUUGUCCGGGAUCCUGUGGCUUCAAUGCCUACUGCAGCGUGGUGAACCACUCGCCCGUCUGCUCUUGC .......(((((...(((.....)))..)))))...((((((((((((((((((.....))))))))).(((......)))....)))))((((....))))..........)))).... ( -48.10) >DroPer_CAF1 39532 120 - 1 AAUUCCGAGUGUCCUGCCAAUCUGGCGUGCAUCAAUGAGCGCUGCACAGACCCUUGUCCGGGGUCAUGCGGCUUCAAUGCCUACUGCAGUGUCGUUGGCCACUCGCCCAUCUGUUCGUGC .....((((((....(((.....))).....(((((((.(((((((..((((((.....))))))....(((......)))...)))))))))))))).))))))............... ( -44.80) >consensus AACACCGAGUGUCCUGCCAAUCUGGCCUGCAUCAAUGAGCGCUGCAAGGAUCCUUGUCCGGGAUCCUGUGGCUUCAAUGCCUACUGCAGUGUCGUAAACCACUCGCCCAUCUGCACCUGC ......(((((...((((.....))))...........(((((((...((((((.....))))))....(((......)))....))))))).......)))))................ (-32.46 = -31.30 + -1.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:02 2006