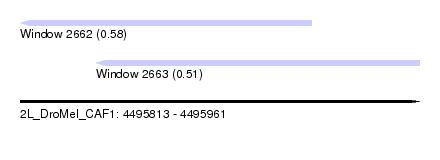
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,495,813 – 4,495,961 |
| Length | 148 |
| Max. P | 0.580233 |

| Location | 4,495,813 – 4,495,921 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 73.37 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -17.87 |
| Energy contribution | -17.57 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4495813 108 - 22407834 GCAUCAGCACAGAGCUUAUUGCUCAUGUCAUCCUGGUUAUACAGGUGAUGCCUUCAUGCGUUGCCAGUCCUUGCCUUCGCCCCAACCGAUUAGGGACUCACCAGUAAU ((....))(((((((.....)))).)))....(((((......((..((((......))))..))((((((((...(((.......))).)))))))).))))).... ( -34.20) >DroVir_CAF1 10251 99 - 1 ACUACAUCACCGAGUUUAUUGCUCUUGCAUCCCCGGCCACACUGGCGAUGCAUUGGUGCGUUGUCGACCCAUACCUC------AGCCCGUUGUGG---CACGCGAACC ......(((((((((.....)))).((((((.((((.....)))).))))))..)))((((....((........))------.(((......))---)))))))... ( -28.40) >DroGri_CAF1 12088 99 - 1 UCUACAUCACCGGGCCCAUUGCUCCUGUAUCCCCAGCCACACUGGCGAUCCCCUUGUGCGUUGUCAGCCCAUACCAC------AACCUGUUGUGG---CCCGCGAACC ......((...((((....(((....)))....((((((((..((.....))..)))).))))...))))...((((------((....))))))---.....))... ( -25.00) >DroSec_CAF1 10295 108 - 1 GCAUCAGCACAGAGCUUAUUGCUCGUGUCAUCCUGGUUAUACAGGUGAUGCCUUCAUGCGCUGCCAGCCCUUGCCUCCGCCCCAACUGAUUAGGGACUCAGCAGUAAU ((((((((((.((((.....))))))))...((((......))))))))))........(((((.((((((....((.(......).))..)))).))..)))))... ( -34.60) >DroEre_CAF1 11035 108 - 1 GCAUCAGCACAGAGCUUAUUGCUCGUGUCAUCCUGGCUAUACAGGUGAUGCCUUCAUGCGCUGCCAGUUCUUGCCUCCGCCCCAACCGAUUAGGGAUUCGCCAGUAAU ((((((((((.((((.....))))))))...((((......))))))))))........(((((.((((((((..((.(......).)).)))))))).).))))... ( -30.80) >DroYak_CAF1 3634 108 - 1 GCAUCAGCACAGAGCUUAUUGCUCGUGUCAUCCUGGUUAUACAGGUGAUGCCUUCAUGCGUUGCCAACCCUUGCCUCCGCCCCAACCGAUUAGGGAUUCACCAGUAAU ......((((.((((.....))))))))....(((((......((..((((......))))..))..((((....((.(......).))..))))....))))).... ( -30.50) >consensus GCAUCAGCACAGAGCUUAUUGCUCGUGUCAUCCUGGCUAUACAGGUGAUGCCUUCAUGCGUUGCCAGCCCUUGCCUCCGCCCCAACCGAUUAGGGA_UCACCAGUAAU ......((...((((.....))))......((((((((.....((((((((......)))))))).......((....)).......))))))))........))... (-17.87 = -17.57 + -0.30)

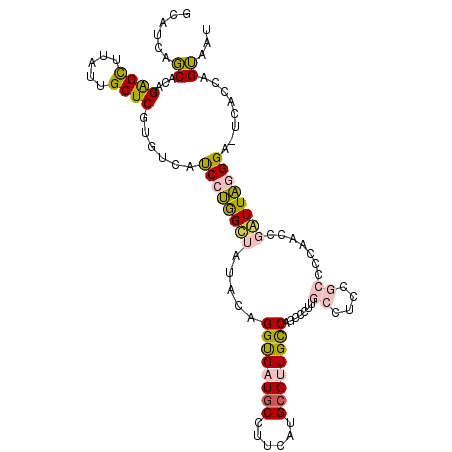
| Location | 4,495,841 – 4,495,961 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.57 |
| Mean single sequence MFE | -39.97 |
| Consensus MFE | -21.19 |
| Energy contribution | -21.45 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.53 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4495841 120 - 22407834 CUUGCGCGGGUGCUUGUGGUCAGAACGCUAUUUGUCGAGCGCAUCAGCACAGAGCUUAUUGCUCAUGUCAUCCUGGUUAUACAGGUGAUGCCUUCAUGCGUUGCCAGUCCUUGCCUUCGC ...(((.((((.....((((...(((((....((..(((.(((((((.(((((((.....)))).))))..((((......))))))))))))))).)))))))))......)))).))) ( -39.40) >DroVir_CAF1 10273 117 - 1 CCUGUCCUGGUAGAUGUGGAUACAAUGCCAUCUGCCAAACACUACAUCACCGAGUUUAUUGCUCUUGCAUCCCCGGCCACACUGGCGAUGCAUUGGUGCGUUGUCGACCCAUACCUC--- ...(((.(((((((((.(.((...)).))))))))))......(((.((((((((.....)))).((((((.((((.....)))).))))))..))))...))).))).........--- ( -37.30) >DroPse_CAF1 11611 120 - 1 CGUGUCCAGGCGCUUGCGGGCAGAAUGCCAUUUGUCAGGCGCAUCAACACCGAGCCCUGUGCUCCUGUCCGCCCGGCUACACCGGCGAUGCCUUCACCCGAUGCCUGCCAUUACCUGCUC ......((((.....(((((((....(((........)))(((((......((((.....))))......(((.((.....))))))))))..........))))))).....))))... ( -39.50) >DroGri_CAF1 12110 117 - 1 CAUGCCCAGGUGCCUGCGGAUACAAUGCAAUUUGCAAGGCUCUACAUCACCGGGCCCAUUGCUCCUGUAUCCCCAGCCACACUGGCGAUCCCCUUGUGCGUUGUCAGCCCAUACCAC--- ..((((.(((((.(((.(((((((..(((((..((..((..........))..))..)))))...))))))).))).))).)))))).......((((.((.....)).))))....--- ( -35.40) >DroSec_CAF1 10323 120 - 1 CUUGUGCGGGUGCUUGUGGUCAGAACGCCAUUUGUCGAGCGCAUCAGCACAGAGCUUAUUGCUCGUGUCAUCCUGGUUAUACAGGUGAUGCCUUCAUGCGCUGCCAGCCCUUGCCUCCGC .....(((((((((((((((......)))).....)))))))..((((.((((((.....))))((((((.((((......)))))))))).....)).)))).............)))) ( -42.70) >DroYak_CAF1 3662 120 - 1 CUUGCGCGGGUGCUUGUGGUCAGAACGCUAUUUGUCGAGCGCAUCAGCACAGAGCUUAUUGCUCGUGUCAUCCUGGUUAUACAGGUGAUGCCUUCAUGCGUUGCCAACCCUUGCCUCCGC .....(((((.((..(.(((..((((((....((..(((.((((((((((.((((.....))))))))...((((......))))))))))))))).))))).)..))))..)).))))) ( -45.50) >consensus CUUGCCCGGGUGCUUGUGGUCAGAACGCCAUUUGUCAAGCGCAUCAGCACAGAGCUUAUUGCUCCUGUCAUCCCGGCUACACAGGCGAUGCCUUCAUGCGUUGCCAGCCCUUACCUCCGC ..(((....(((((((....((((......)))).)))))))....)))..((((.....))))...................((((((((......))))))))............... (-21.19 = -21.45 + 0.26)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:53 2006