
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,485,362 – 4,485,507 |
| Length | 145 |
| Max. P | 0.999915 |

| Location | 4,485,362 – 4,485,473 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.16 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -13.44 |
| Energy contribution | -15.30 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.58 |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.998470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
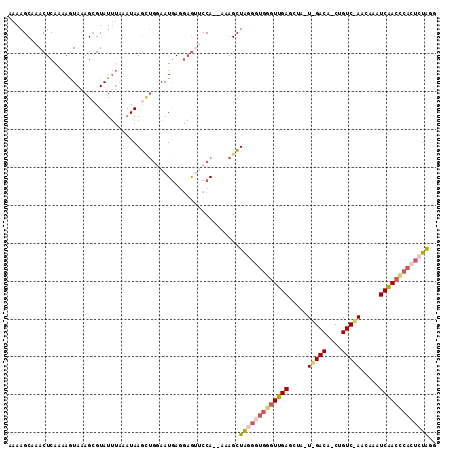
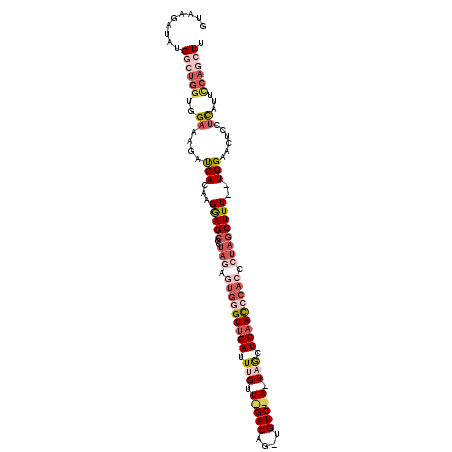
>2L_DroMel_CAF1 4485362 111 + 22407834 AAAAGCAAACUCAAAAGUAAAGCGUAUUUAAAUAAGUUGGAAUAAGGAGUACCA--AAAGCUGGGGUGGGUUGAGCUAUUGGACAGCUGUCUAACAAAUCAACCCACUCUAAG ........((((........(((.(((....))).)))........))))....--.....(((((((((((((....((((((....))))))....))))))))))))).. ( -32.59) >DroSec_CAF1 670 89 + 1 AAAAGCAAACUCAAAAAUAAAGCGUAUUUAAUUAAU---UAAUGAGGAGUCCCA--AAAGAUAGGGUGGGUUGAUCU-------------------AAUCGACUCACUCUAGG .........((((..(((.((..........)).))---)..))))........--.....((((((((((((((..-------------------.)))))))))))))).. ( -19.10) >DroSim_CAF1 675 87 + 1 AAAAGCAAACUCAAAAAUAAAGCGUAUUUAAAUAAGCUGAAAUGACGAGUUCCA--AAAGCUAG-----GUUGAGCU-------------------AAUCAACCCACUCUAGG ...(((.(((((........(((.(((....))).)))........)))))...--...))).(-----(((((...-------------------..))))))......... ( -14.49) >DroEre_CAF1 676 107 + 1 AAAAGCAAACUCAAAAGUAAAGGGUAU---AAUAAGCUGGAAUGA---AAUCCACCAAAGCUAUGGUGGAUUGAGUUAAUAGACAGCUGUCUAACAAAUCAACCCACUUUAAG .............(((((...((((..---..........(((..---(((((((((......)))))))))..)))..(((((....)))))........)))))))))... ( -26.90) >DroYak_CAF1 659 111 + 1 AAAAGCAAACUCAAAAGUAAAGCGUAUUUAAAUAAGCUGGAAUGAGGAGAUCCA--AAAGCUAUGUUGGGUUGAGUUAAUAGACAUCUGUCCAACAAAUCAACCCACUCUGGA ...(((....(((....(..(((.(((....))).)))..).)))((....)).--...)))....((((((((.......(((....))).......))))))))....... ( -21.94) >consensus AAAAGCAAACUCAAAAGUAAAGCGUAUUUAAAUAAGCUGGAAUGAGGAGUUCCA__AAAGCUAGGGUGGGUUGAGCUA_U_GACA_CUGUC_AACAAAUCAACCCACUCUAGG .............................................................(((((((((((((.....(((((....))))).....))))))))))))).. (-13.44 = -15.30 + 1.86)


| Location | 4,485,362 – 4,485,473 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.16 |
| Mean single sequence MFE | -23.55 |
| Consensus MFE | -9.84 |
| Energy contribution | -11.76 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.42 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
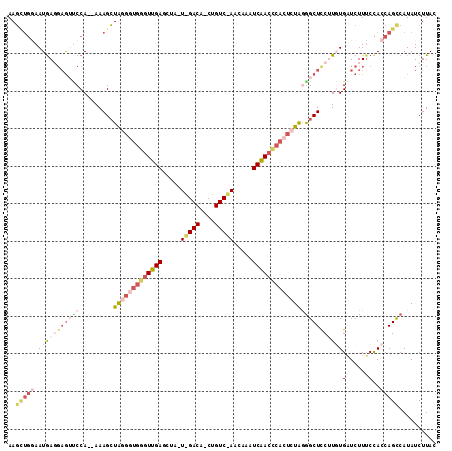
>2L_DroMel_CAF1 4485362 111 - 22407834 CUUAGAGUGGGUUGAUUUGUUAGACAGCUGUCCAAUAGCUCAACCCACCCCAGCUUU--UGGUACUCCUUAUUCCAACUUAUUUAAAUACGCUUUACUUUUGAGUUUGCUUUU .((((((((((((((.(((((.(((....))).))))).)))))))))........(--(((...........))))....)))))....((...(((....)))..)).... ( -25.80) >DroSec_CAF1 670 89 - 1 CCUAGAGUGAGUCGAUU-------------------AGAUCAACCCACCCUAUCUUU--UGGGACUCCUCAUUA---AUUAAUUAAAUACGCUUUAUUUUUGAGUUUGCUUUU ....(((.((((((((.-------------------..))).......((.......--.))))))))))....---.............((((.......))))........ ( -13.40) >DroSim_CAF1 675 87 - 1 CCUAGAGUGGGUUGAUU-------------------AGCUCAAC-----CUAGCUUU--UGGAACUCGUCAUUUCAGCUUAUUUAAAUACGCUUUAUUUUUGAGUUUGCUUUU ((.((((((((((((..-------------------...)))))-----))).))))--.))((((((.......(((.(((....))).))).......))))))....... ( -21.34) >DroEre_CAF1 676 107 - 1 CUUAAAGUGGGUUGAUUUGUUAGACAGCUGUCUAUUAACUCAAUCCACCAUAGCUUUGGUGGAUU---UCAUUCCAGCUUAUU---AUACCCUUUACUUUUGAGUUUGCUUUU (((((((((((((((.(((.(((((....))))).))).))))))))).........(.((((..---....)))).).....---............))))))......... ( -28.00) >DroYak_CAF1 659 111 - 1 UCCAGAGUGGGUUGAUUUGUUGGACAGAUGUCUAUUAACUCAACCCAACAUAGCUUU--UGGAUCUCCUCAUUCCAGCUUAUUUAAAUACGCUUUACUUUUGAGUUUGCUUUU (((((((((((((((.(((.(((((....))))).))).)))))))).......)))--))))............(((..(((((((...........)))))))..)))... ( -29.21) >consensus CCUAGAGUGGGUUGAUUUGUU_GACAG_UGUC_A_UAGCUCAACCCACCCUAGCUUU__UGGAACUCCUCAUUCCAGCUUAUUUAAAUACGCUUUACUUUUGAGUUUGCUUUU ..(((((((((((((.(((.(((((....))))).))).))))))).....((((....(((......)))....))))...........))))))................. ( -9.84 = -11.76 + 1.92)


| Location | 4,485,395 – 4,485,507 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 72.86 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -17.16 |
| Energy contribution | -22.34 |
| Covariance contribution | 5.18 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.54 |
| SVM decision value | 4.53 |
| SVM RNA-class probability | 0.999915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
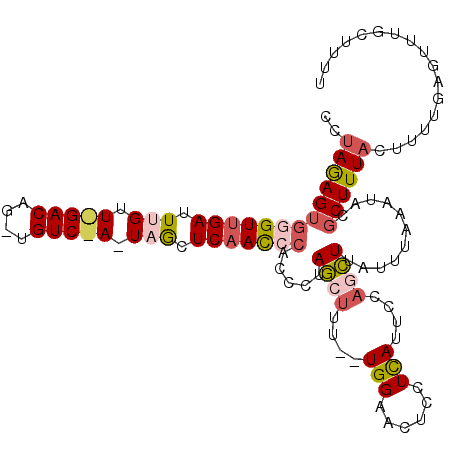
>2L_DroMel_CAF1 4485395 112 + 22407834 AAGUUGGAAUAAGGAGUACCA--AAAGCUGGGGUGGGUUGAGCUAUUGGACAGCUGUCUAACAAAUCAACCCACUCUAAGGCUUCUUUUGGUCUUUCCACCAGCCAUAUCUUAC ..(((((.....((((.((((--((((((((((((((((((....((((((....))))))....))))))))))))..))))...))))))..)))).))))).......... ( -44.40) >DroSec_CAF1 703 90 + 1 AAU---UAAUGAGGAGUCCCA--AAAGAUAGGGUGGGUUGAUCU-------------------AAUCGACUCACUCUAGGGCUCCAUGUGAUCUUACUACCAGCCAUAUCUUAC ...---......(((((((..--.....((((((((((((((..-------------------.)))))))))))))))))))))(((((..............)))))..... ( -29.15) >DroSim_CAF1 708 88 + 1 AAGCUGAAAUGACGAGUUCCA--AAAGCUAG-----GUUGAGCU-------------------AAUCAACCCACUCUAGGGCUCCAUGUGAUCUUUCCACCAGCCAUAUCUUAC ..((((.......((((((..--..((...(-----(((((...-------------------..))))))..))...))))))...(((.......))))))).......... ( -19.00) >DroEre_CAF1 706 111 + 1 AAGCUGGAAUGA---AAUCCACCAAAGCUAUGGUGGAUUGAGUUAAUAGACAGCUGUCUAACAAAUCAACCCACUUUAAGGCUCCUUGUGAUCUUUCCACCAGUCAUAUCUUAC ..(((((...((---((..(((...((((..(((((.((((.....(((((....))))).....)))).)))))....))))....)))...))))..))))).......... ( -29.00) >DroYak_CAF1 692 112 + 1 AAGCUGGAAUGAGGAGAUCCA--AAAGCUAUGUUGGGUUGAGUUAAUAGACAUCUGUCCAACAAAUCAACCCACUCUGGAGCUCCUUUUGAUCUUUCCACCAGCCAUAUCUUAC ..(((((..((..((((((.(--((((((..(.((((((((.......(((....))).......))))))))..)...))))..))).))))))..))))))).......... ( -36.84) >consensus AAGCUGGAAUGAGGAGUUCCA__AAAGCUAGGGUGGGUUGAGCUA_U_GACA_CUGUC_AACAAAUCAACCCACUCUAGGGCUCCUUGUGAUCUUUCCACCAGCCAUAUCUUAC ..(((((.(((((((((((.........(((((((((((((.....(((((....))))).....))))))))))))))))))))))))((....))..))))).......... (-17.16 = -22.34 + 5.18)

| Location | 4,485,395 – 4,485,507 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.86 |
| Mean single sequence MFE | -31.88 |
| Consensus MFE | -16.06 |
| Energy contribution | -20.22 |
| Covariance contribution | 4.16 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
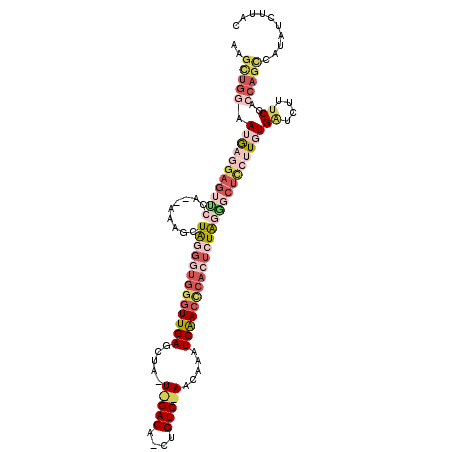
>2L_DroMel_CAF1 4485395 112 - 22407834 GUAAGAUAUGGCUGGUGGAAAGACCAAAAGAAGCCUUAGAGUGGGUUGAUUUGUUAGACAGCUGUCCAAUAGCUCAACCCACCCCAGCUUU--UGGUACUCCUUAUUCCAACUU ........(((.....(((...((((((((....((..(.(((((((((.(((((.(((....))).))))).))))))))))..))))))--))))..))).....))).... ( -36.20) >DroSec_CAF1 703 90 - 1 GUAAGAUAUGGCUGGUAGUAAGAUCACAUGGAGCCCUAGAGUGAGUCGAUU-------------------AGAUCAACCCACCCUAUCUUU--UGGGACUCCUCAUUA---AUU ....(((...((.....))...)))....(((((((.((((((.((.(((.-------------------..))).)).)))....)))..--.))).))))......---... ( -18.80) >DroSim_CAF1 708 88 - 1 GUAAGAUAUGGCUGGUGGAAAGAUCACAUGGAGCCCUAGAGUGGGUUGAUU-------------------AGCUCAAC-----CUAGCUUU--UGGAACUCGUCAUUUCAGCUU ....((.(((((..((((.....))))...(((.((.((((((((((((..-------------------...)))))-----))).))))--.))..)))))))).))..... ( -25.80) >DroEre_CAF1 706 111 - 1 GUAAGAUAUGACUGGUGGAAAGAUCACAAGGAGCCUUAAAGUGGGUUGAUUUGUUAGACAGCUGUCUAUUAACUCAAUCCACCAUAGCUUUGGUGGAUU---UCAUUCCAGCUU ...........((((..((((..((((...((((......(((((((((.(((.(((((....))))).))).)))))))))....))))..)))).))---))...))))... ( -35.40) >DroYak_CAF1 692 112 - 1 GUAAGAUAUGGCUGGUGGAAAGAUCAAAAGGAGCUCCAGAGUGGGUUGAUUUGUUGGACAGAUGUCUAUUAACUCAACCCAACAUAGCUUU--UGGAUCUCCUCAUUCCAGCUU .........((((((..((.(((((...(((((((......((((((((.(((.(((((....))))).))).))))))))....))))))--).)))))..))...)))))). ( -43.20) >consensus GUAAGAUAUGGCUGGUGGAAAGAUCACAAGGAGCCCUAGAGUGGGUUGAUUUGUU_GACAG_UGUC_A_UAGCUCAACCCACCCUAGCUUU__UGGAACUCCUCAUUCCAGCUU .........((((((..((....(((...(((((..(((.(((((((((.(((.(((((....))))).))).))))))))).))))))))..)))......))...)))))). (-16.06 = -20.22 + 4.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:49 2006