
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,479,826 – 4,479,934 |
| Length | 108 |
| Max. P | 0.500000 |

| Location | 4,479,826 – 4,479,934 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -20.82 |
| Energy contribution | -21.02 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4479826 108 + 22407834 A----UUUC------AGGACUACUCACUGUUGGCAUCAUUGAGACCUUGACCGCUGCGCUUCUCGAAUGUCAAAAUGGCAUUUGAUGAGAUUGUGAUCUCUUCGCGCAGUUGACCCUC .----..((------(((....((((.((.......)).))))..)))))(.(((((((...(((((((((.....))))))))).(((((....)))))...))))))).)...... ( -34.00) >DroVir_CAF1 19219 108 + 1 U----GGGU------AGGGGUACUCACUGUUGGCAUCAUUCAGGCCUGAACCGCUGCGUUUCUCGAAUGUCAAUAUGGCAUUGGAAGAGAUAGUUAUCUCCUCACGCAACUGACCCUC .----((((------((.(((.(...(((.((....))..)))....).))).))((((...((.((((((.....)))))).)).(((((....)))))...)))).....)))).. ( -30.60) >DroGri_CAF1 16767 108 + 1 U----GUAU------AGGGGUACUCACUGUUGGCAUCAUUCAGGCCAGGACCGCUGCGUUUCUCGAAUGUCAGGAUGGCAUUAGAGGAGAUGGUGAUCUCCUCGCGCAGUUGACCCUC .----....------.(((((........(((((.........)))))..(.(((((((......((((((.....)))))).((((((((....))))))))))))))).)))))). ( -40.20) >DroWil_CAF1 19073 112 + 1 AGUCACUAA------CGAAUCACUUACUGUUGGCAUCAUUCAGUCCUGGACCACUGCGUUUCUCAAAUGUCAGUAUGGCAUUCGAUGAGAUGGUAAUCUCUUCACGCAGUUGUCCCUC .....((((------((..........))))))..............((((.(((((((......((((((.....)))))).((.(((((....))))).))))))))).))))... ( -31.10) >DroMoj_CAF1 18974 114 + 1 U----AGGUAGUAGUAGGGGUACUCACUGUUGGCAUCAUUCAGGCCUGGACCGCUGCGUUUCUCGAAUGUCAAUAUGGCAUUGGAUGAGAUGGUGAUCUCCUCACGCAGCUGACCCUC .----...........(((((..........(((.........)))......((((((((((((.((((((.....)))))).)).))))).((((.....)))))))))..))))). ( -34.90) >DroAna_CAF1 19195 103 + 1 A----UUU-----------AAACUCACUAUUGGCAUCAUUUAGUCCCGGUCCACUGCGCUUCUCGAACGUCAGGAUGGCAUUCGACGAAAUGGUGAUCUCUUCGCGCAGCUGACCCUC .----...-----------............(((........)))..((((..((((((((((((((.(((.....))).))))).))).....((.....))))))))..))))... ( -27.60) >consensus A____GUAU______AGGGGUACUCACUGUUGGCAUCAUUCAGGCCUGGACCGCUGCGUUUCUCGAAUGUCAAUAUGGCAUUCGAUGAGAUGGUGAUCUCCUCACGCAGCUGACCCUC ..........................(((.((....))..)))....((.(.(((((((......((((((.....)))))).((.(((((....))))).))))))))).).))... (-20.82 = -21.02 + 0.20)

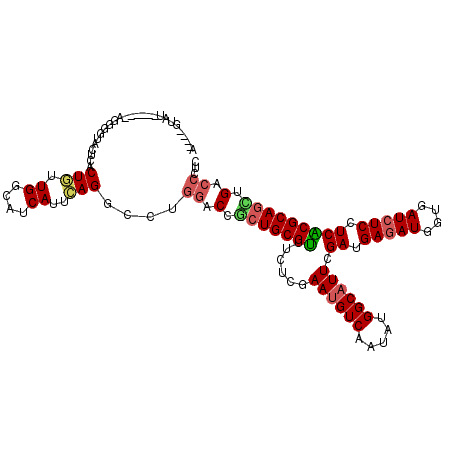
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:44 2006