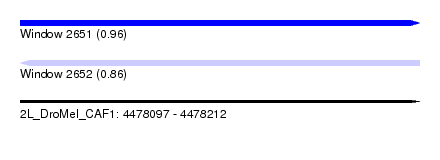
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,478,097 – 4,478,212 |
| Length | 115 |
| Max. P | 0.959246 |

| Location | 4,478,097 – 4,478,212 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.19 |
| Mean single sequence MFE | -45.46 |
| Consensus MFE | -39.34 |
| Energy contribution | -39.46 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
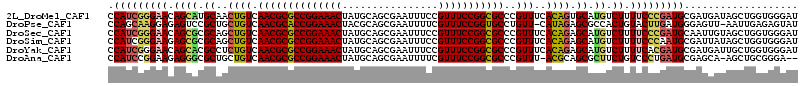
>2L_DroMel_CAF1 4478097 115 + 22407834 AUCCCACCAGCUAUCAUCGCAUCGGGAAAAGACAUGCACUGUGAAACGGGCGCCGGAAACGGAAAUUCGCUGCAUAGUUUCCGGCGCGUUGACAGUUGCAUGCUGUUCCCGAUGG ...................(((((((((.((.((((((((((.((....(((((((((((................))))))))))).)).)))).)))))))).))))))))). ( -53.19) >DroPse_CAF1 37126 113 + 1 AUACUCUCAAUU-AACUCCCAUCAAGUACAGUGGCGCUCUAUG-AACAGGCACCGGAAACGAAAAUUCGCUGCGUAGUUUCCGGUGCGUUGACAGCAGCGGACUCUCCUUGCUGG ..........((-(((...(((..((..(....)..))..)))-.....(((((((((((................))))))))))))))))((((((.((.....)))))))). ( -35.19) >DroSec_CAF1 14109 115 + 1 AUCCCACCAGCUACAAUUGCAUCGGGAAAAGACAUGCUCUGUGAAACGGGCGCCGGAAACGGAAAUUCGCUGCAUAGUUUCCGGCGCGUUGACAGCUGCGCGCUGUUCCCGAUGG ...................(((((((((.((.(.(((.((((.((....(((((((((((................))))))))))).)).))))..))).))).))))))))). ( -46.19) >DroSim_CAF1 14106 115 + 1 AUCCCACCAGCUAUAAUCGCAUUGGGAAAAGACAUGCUCUGUGAAACGGGCGCCGGAAACGGAAAUUCGCUGCAUAGUUUCCGGCGCGUUGACAGCUGCGCGCUCUUCCCGAUGG ...................(((((((((.((.(.(((.((((.((....(((((((((((................))))))))))).)).))))..))).))).))))))))). ( -44.09) >DroYak_CAF1 14487 115 + 1 AUCCCACCAGCAAUCAUCGCAUCGUGAAAAGACAUGCUCUGUGAAACGGGCGCCGGAAACGGAAAUUCGCUGCAUAGUUUCCGGCGCGUUGACAGAGGCGUGCUGUUCCCGAUGG ...................(((((.(((.((.((((((((((.((....(((((((((((................))))))))))).)).))))).))))))).))).))))). ( -45.89) >DroAna_CAF1 17542 111 + 1 --UCCCGCAGCU-UGCUCGCAUCAGGGACAGAAGCGCUGCGU-AAACGGGCGCCGGAAACGAAAAUUCGCUGCAUAGUUUCCGGCGCGUUGACAGCAGCGCCCUCUUCCGGAUGG --....((....-.))...((((..(((.(((.(((((((((-.(((..(((((((((((................)))))))))))))).)).)))))))..)))))).)))). ( -48.19) >consensus AUCCCACCAGCUAUAAUCGCAUCGGGAAAAGACAUGCUCUGUGAAACGGGCGCCGGAAACGGAAAUUCGCUGCAUAGUUUCCGGCGCGUUGACAGCUGCGCGCUCUUCCCGAUGG ...................(((((((((.((...((((((((.((....(((((((((((................))))))))))).)).)))).))))..)).))))))))). (-39.34 = -39.46 + 0.12)



| Location | 4,478,097 – 4,478,212 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.19 |
| Mean single sequence MFE | -44.84 |
| Consensus MFE | -34.25 |
| Energy contribution | -34.26 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4478097 115 - 22407834 CCAUCGGGAACAGCAUGCAACUGUCAACGCGCCGGAAACUAUGCAGCGAAUUUCCGUUUCCGGCGCCCGUUUCACAGUGCAUGUCUUUUCCCGAUGCGAUGAUAGCUGGUGGGAU ((((((((((.((((((((.((((.((((((((((((((................)))))))))))..)))..)))))))))).)).)))))...((.......)).)))))... ( -50.99) >DroPse_CAF1 37126 113 - 1 CCAGCAAGGAGAGUCCGCUGCUGUCAACGCACCGGAAACUACGCAGCGAAUUUUCGUUUCCGGUGCCUGUU-CAUAGAGCGCCACUGUACUUGAUGGGAGUU-AAUUGAGAGUAU (((.((((.(.(((.((((.((((.((((((((((((((................)))))))))))..)))-.))))))))..))).).)))).))).....-............ ( -38.09) >DroSec_CAF1 14109 115 - 1 CCAUCGGGAACAGCGCGCAGCUGUCAACGCGCCGGAAACUAUGCAGCGAAUUUCCGUUUCCGGCGCCCGUUUCACAGAGCAUGUCUUUUCCCGAUGCAAUUGUAGCUGGUGGGAU ((((((((((.((((.((..((((.((((((((((((((................)))))))))))..)))..)))).)).)).)).)))))..(((....)))...)))))... ( -42.79) >DroSim_CAF1 14106 115 - 1 CCAUCGGGAAGAGCGCGCAGCUGUCAACGCGCCGGAAACUAUGCAGCGAAUUUCCGUUUCCGGCGCCCGUUUCACAGAGCAUGUCUUUUCCCAAUGCGAUUAUAGCUGGUGGGAU (((((((((((((((.((..((((.((((((((((((((................)))))))))))..)))..)))).)).)).))))))))...((.......)).)))))... ( -46.99) >DroYak_CAF1 14487 115 - 1 CCAUCGGGAACAGCACGCCUCUGUCAACGCGCCGGAAACUAUGCAGCGAAUUUCCGUUUCCGGCGCCCGUUUCACAGAGCAUGUCUUUUCACGAUGCGAUGAUUGCUGGUGGGAU (((((((.((((...(((((((((.((((((((((((((................)))))))))))..)))..))))))...(((.......)))))).)).)).)))))))... ( -42.39) >DroAna_CAF1 17542 111 - 1 CCAUCCGGAAGAGGGCGCUGCUGUCAACGCGCCGGAAACUAUGCAGCGAAUUUUCGUUUCCGGCGCCCGUUU-ACGCAGCGCUUCUGUCCCUGAUGCGAGCA-AGCUGCGGGA-- ...((((((.(..((((((((.((.((((((((((((((................)))))))))))..))).-))))))))))..).)))....(((.((..-..))))))))-- ( -47.79) >consensus CCAUCGGGAACAGCGCGCAGCUGUCAACGCGCCGGAAACUAUGCAGCGAAUUUCCGUUUCCGGCGCCCGUUUCACAGAGCAUGUCUUUUCCCGAUGCGAUUAUAGCUGGUGGGAU .(((((((((.((((.((..((((.((((((((((((((................)))))))))))..)))..)))).)).)).)).)))))))))................... (-34.25 = -34.26 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:42 2006