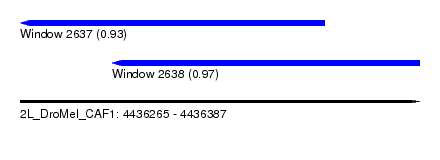
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,436,265 – 4,436,387 |
| Length | 122 |
| Max. P | 0.968827 |

| Location | 4,436,265 – 4,436,358 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -22.26 |
| Consensus MFE | -16.88 |
| Energy contribution | -17.04 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4436265 93 - 22407834 GCGACACAAAAAGUGGGCAAAAAAAAAAAAUGAAAGAAAAAGUGUAAUAUUCACUUUUUAUGCUGAGUCCGGCGUUUCUCGCAUGCGACUAAA (((((((.....)))................((((.((((((((.......)))))))).(((((....)))))))))))))........... ( -19.50) >DroSec_CAF1 41737 86 - 1 GCGCCACAAAAAGUGGGCAAAAAAAGGA------AG-AAAAGUGUAAUAUUCACUUUUUAUGCUGAGUCCGGCGUUUCUCGCAUGCGACUAAA ((.((((.....))))))......((.(------.(-(((((((.......)))))))).).)).(((((((((.....))).)).))))... ( -24.20) >DroSim_CAF1 45956 86 - 1 GCGCCACAAAAAGUGGGCAAAAAAAGGA------AG-AAAAGUGUAAUAUUCACUUUUUAUGCUGAGCCCGGCGUUUCUCGCAUGCGACUAAA (((((((.....))((((......((.(------.(-(((((((.......)))))))).).))..)))))))))...(((....)))..... ( -24.10) >DroEre_CAF1 43936 82 - 1 GCGCCACAAAAAGUGGGCAAAAAAA----------U-AAAAGUGUAAUAUUCACUUUUUAUGCUGAGGCUGGCGCUUCUCGCAUGCGACUAAA ((.((((.....)))))).......----------.-(((((((.......)))))))(((((.(((((....)))))..)))))........ ( -23.00) >DroYak_CAF1 42627 82 - 1 GCGCCACAAAAAGUGGGCAAAAAAA----------U-AAAAGUGUAAUAUUCACUUUUUAUGCUGAGGCUGGCGUUUCUCGCAUGCGACUAAA ((.((((.....)))))).......----------.-(((((((.......)))))))(((((.(((((....)))))..)))))........ ( -20.50) >consensus GCGCCACAAAAAGUGGGCAAAAAAA__A______AG_AAAAGUGUAAUAUUCACUUUUUAUGCUGAGGCCGGCGUUUCUCGCAUGCGACUAAA (((((((.....))))((...................(((((((.......))))))).((((((....)))))).....)).)))....... (-16.88 = -17.04 + 0.16)


| Location | 4,436,293 – 4,436,387 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 90.82 |
| Mean single sequence MFE | -18.74 |
| Consensus MFE | -15.56 |
| Energy contribution | -15.76 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4436293 94 - 22407834 GGCUUUAACUUGGCCCUCUACCCUAAAAAGCGACACAAAAAGUGGGCAAAAAAAAAAAAUGAAAGAAAAAGUGUAAUAUUCACUUUUUAUGCUG ((((.......)))).............((((.(((.....))).....................((((((((.......)))))))).)))). ( -16.10) >DroSec_CAF1 41765 87 - 1 GGCUUUAACUUGGCCCUCUACCCCAAAAAGCGCCACAAAAAGUGGGCAAAAAAAGGA------AG-AAAAGUGUAAUAUUCACUUUUUAUGCUG ((((.......))))..............((.((((.....))))))......((.(------.(-(((((((.......)))))))).).)). ( -20.70) >DroSim_CAF1 45984 87 - 1 GGCUUUAACUUGGCCCUCUACCCCAAAAAGCGCCACAAAAAGUGGGCAAAAAAAGGA------AG-AAAAGUGUAAUAUUCACUUUUUAUGCUG ((((.......))))..............((.((((.....))))))......((.(------.(-(((((((.......)))))))).).)). ( -20.70) >DroEre_CAF1 43964 82 - 1 GGCUUUAACUUGGCCCUCUACC-CAAAAUGCGCCACAAAAAGUGGGCAAAAAAA----------U-AAAAGUGUAAUAUUCACUUUUUAUGCUG ((((.......)))).......-.....(((.((((.....)))))))......----------.-(((((((.......)))))))....... ( -18.10) >DroYak_CAF1 42655 82 - 1 GGCUUUAACUUGGCCCUCUACC-CAAAAUGCGCCACAAAAAGUGGGCAAAAAAA----------U-AAAAGUGUAAUAUUCACUUUUUAUGCUG ((((.......)))).......-.....(((.((((.....)))))))......----------.-(((((((.......)))))))....... ( -18.10) >consensus GGCUUUAACUUGGCCCUCUACCCCAAAAAGCGCCACAAAAAGUGGGCAAAAAAA__A______AG_AAAAGUGUAAUAUUCACUUUUUAUGCUG ((((.......))))..............((.((((.....))))))...................(((((((.......)))))))....... (-15.56 = -15.76 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:27 2006