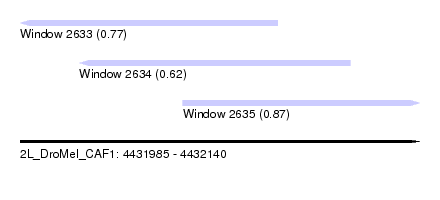
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,431,985 – 4,432,140 |
| Length | 155 |
| Max. P | 0.868846 |

| Location | 4,431,985 – 4,432,085 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 88.36 |
| Mean single sequence MFE | -23.54 |
| Consensus MFE | -19.79 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4431985 100 - 22407834 UGUUCGCCACCUCACCUUUCCACCAGCCCACCUGCUCACCUGGGCAACUGGGCAUUUGGGCACACACUCGCAUACAAAUUGCUUCCUGUCCCAGUCUCCC .........................(((((..(((((....)))))..)))))..(((((.(((.....(((.......)))....))))))))...... ( -26.60) >DroSec_CAF1 37497 92 - 1 UG--------CUCACCUGUUCGCCACCUCACCUGCCCACCUGGGCAACUGGGCAUUUGGACACACACUCGCAUACAAAUUGCUUCCUGUCCCAGUCUCCC ..--------.....(((...(((....((..(((((....)))))..)))))....(((((.......(((.......)))....))))))))...... ( -21.30) >DroSim_CAF1 41081 92 - 1 UG--------CUCACCUGUUCGCCACCUCUCCUGCCCACCGGGGCAACUGGGCAUUUGGGCACACACUCGCAUACAAAUUGCUUCCUGUCCCAGUCUCCC ((--------((((...(((.(((.((.............)))))))))))))).(((((.(((.....(((.......)))....))))))))...... ( -22.72) >consensus UG________CUCACCUGUUCGCCACCUCACCUGCCCACCUGGGCAACUGGGCAUUUGGGCACACACUCGCAUACAAAUUGCUUCCUGUCCCAGUCUCCC ................................(((((....)))))((((((((...(((......)))(((.......)))....)).))))))..... (-19.79 = -19.90 + 0.11)

| Location | 4,432,008 – 4,432,113 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.39 |
| Mean single sequence MFE | -23.07 |
| Consensus MFE | -8.59 |
| Energy contribution | -11.35 |
| Covariance contribution | 2.76 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4432008 105 - 22407834 CAGGUGCUCACCAGUUCACCAGCUCACCUGUUCGCCACCUCACCUUUCCACCAGCCCACCUGCUCACCUGGGCAACUGGGCAUUUGGGCACACACUCGCAUACAA ...(((((((.........(((.....))).......................(((((..(((((....)))))..)))))...))))))).............. ( -27.90) >DroSec_CAF1 37520 97 - 1 CAGGUGCUCACCAAUUCACCAGUUCACCUG--------CUCACCUGUUCGCCACCUCACCUGCCCACCUGGGCAACUGGGCAUUUGGACACACACUCGCAUACAA ((((((..(............)..))))))--------......((((((....((((..(((((....)))))..))))....))))))............... ( -24.00) >DroSim_CAF1 41104 97 - 1 CAGGUGCUCACCAGUUCACCAGUUCACCUG--------CUCACCUGUUCGCCACCUCUCCUGCCCACCGGGGCAACUGGGCAUUUGGGCACACACUCGCAUACAA ...(((((((.(((.....(((.....)))--------.....)))...(((........(((((....)))))....)))...))))))).............. ( -25.20) >DroEre_CAF1 39707 89 - 1 CAGGUGAGCACCAGUUCACCAGCGCACCAG--------CUCACCUGUUCGCCACCUCACCUUUCCACCUGCCCACC--------UGGGCACACACUCGCAUACAA (((((((((....((......))......)--------))))))))......................(((((...--------.)))))............... ( -24.00) >DroYak_CAF1 38465 89 - 1 CAGGUGCUCACCAGUUCACCAGCGCACCAGCUCACCAGCUCACCUGUUCGCCACCUCACCUUUCCA--------CC--------UGGGCACACACUCGCAUACAA ((((((......((((....(((......)))....))))))))))...(((..............--------..--------..)))................ ( -14.27) >consensus CAGGUGCUCACCAGUUCACCAGCUCACCUG________CUCACCUGUUCGCCACCUCACCUGCCCACCUGGGCAACUGGGCAUUUGGGCACACACUCGCAUACAA ((((((..................))))))..............((((((....(((...(((((....)))))...)))....))))))............... ( -8.59 = -11.35 + 2.76)

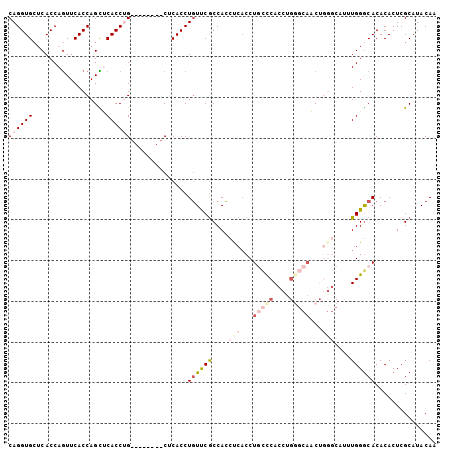
| Location | 4,432,048 – 4,432,140 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 71.96 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -9.92 |
| Energy contribution | -10.98 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4432048 92 + 22407834 GAGCAGGUGG-GCUGGUGGAAAGGUG-AGGUGGCGAA-CAGGUGAGCUGGUGAACUGGUGAGCACCUGGGGU-CAGCGAGCGGGCGUCAC---AAGAAU ......((((-(((.((.........-.(.((((...-((((((.((..(....)..))...))))))..))-)).)..)).))).))))---...... ( -31.60) >DroSec_CAF1 37560 84 + 1 GGGCAGGUGA-GGUGGCGAACAGGUG-AG---------CAGGUGAACUGGUGAAUUGGUGAGCACCUGGGGU-CAGCGGGCGGGCGUCAC---AAGAAU ..........-.((((((..(.....-..---------((((((.((..(....)..))...))))))..((-(....))))..))))))---...... ( -25.70) >DroSim_CAF1 41144 84 + 1 GGGCAGGAGA-GGUGGCGAACAGGUG-AG---------CAGGUGAACUGGUGAACUGGUGAGCACCUGGGGU-CAGCGGGCGGGCGUCAC---CAGAAU ..........-(((((((..(.....-..---------((((((.((..(....)..))...))))))..((-(....))))..))))))---)..... ( -33.00) >DroEre_CAF1 39739 84 + 1 GGAAAGGUGA-GGUGGCGAACAGGUG-AG---------CUGGUGCGCUGGUGAACUGGUGCUCACCUGGGGU-CAGCGGGAGGGCGUCAC---AAAAAU ..........-.(((((((.((((((-((---------(......((..(....)..)))))))))))...)-).((......)))))))---...... ( -31.40) >DroYak_CAF1 38489 92 + 1 GGAAAGGUGA-GGUGGCGAACAGGUG-AGCUGGUGAG-CUGGUGCGCUGGUGAACUGGUGAGCACCUGGGGU-CAGCGGGCAGGCGUCAC---AAAAAU ..........-.((((((..((((((-.((..((..(-(..(....)..))..))..))...))))))..((-(....)))...))))))---...... ( -34.50) >DroPer_CAF1 48833 89 + 1 GGAAAGGUAUAUGUAUUGA-----UGUAUGUGGUAUGUAUGGUAUGUCUG-CAUCUGGUGGGC----GGAGUAAAGCGGGCAGGCAUUCCCCUAAAAAU ((((...((((((((((..-----.......))))))))))((.((((((-(.((((.....)----))).....))))))).)).))))......... ( -20.90) >consensus GGAAAGGUGA_GGUGGCGAACAGGUG_AG_________CAGGUGAGCUGGUGAACUGGUGAGCACCUGGGGU_CAGCGGGCGGGCGUCAC___AAAAAU ............(((((.......................((((.(((((....)))))...)))).........((......)))))))......... ( -9.92 = -10.98 + 1.06)

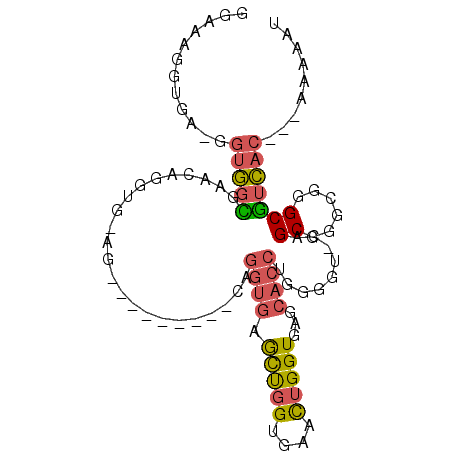
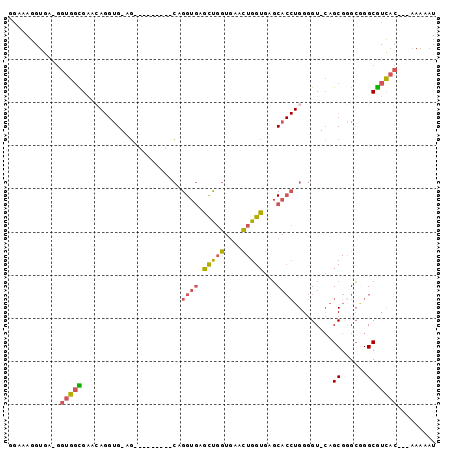
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:25 2006