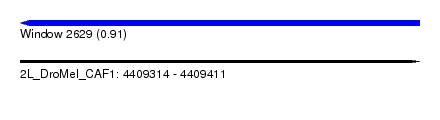
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,409,314 – 4,409,411 |
| Length | 97 |
| Max. P | 0.905130 |

| Location | 4,409,314 – 4,409,411 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.17 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -16.89 |
| Energy contribution | -16.62 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4409314 97 - 22407834 UAA-UUUUA------ACAUAAAUCCCG--UUUUAUAUCUU-CUGCUUGCAGAAAAUGUCUUUAUAGAGAUGGCCAAACUUUCACGACGUGCCUCGUCGUCUGCAGCA ...-.....------..(((((.....--.))))).....-..(((.(((((....(((((....))))).............(((((.....))))))))))))). ( -20.80) >DroSec_CAF1 14995 94 - 1 ----UUUAA------AUAUAUAUCCCG--UUUUAUAUCUU-CUGCUUGCAGAAAAUGUCUUUAUAGAGAUGGCCAAACUUUCACGACGUGCCUCGUCGUCUGCAGCA ----.....------..(((((.....--...)))))...-..(((.(((((....(((((....))))).............(((((.....))))))))))))). ( -20.50) >DroSim_CAF1 16339 94 - 1 ----UUUAA------AUAGAUAUCCCG--UUUUAUAUCUU-CUGCUUGCAGAAAAUGUCUUUAUAGAGAUGGCCAAACUUUCACGACGUGCCUCGUCGUCUGCAGCA ----.....------..((((((....--....)))))).-..(((.(((((....(((((....))))).............(((((.....))))))))))))). ( -22.60) >DroYak_CAF1 15127 87 - 1 U-----------------AAUGUAUUA--AUUGAUACCUU-CUGCUUGCAGAAAAUGUCUUAAUAGAGAUGGCCAAACUUUCACGACGUGCCUCGUCGUCUGCAGCA .-----------------...(((((.--...)))))...-..(((.(((((....(((((....))))).............(((((.....))))))))))))). ( -21.90) >DroAna_CAF1 18823 91 - 1 UAGUUUUAA------AGAGAAAU------AUUUAU---UU-CUCGUUUCAGAAAAUGUUUCAAUAGAGAUGGCCAAACUUUCACGACGUGCCUCGUCGUCUGCAGCA .........------.(((((((------....))---))-)))(((.((((....(((((....))))).............(((((.....))))))))).))). ( -21.50) >DroPer_CAF1 19611 106 - 1 CAA-AUUACAAACGAAUACUAAUUCUAUUGUUGACUCUUUAUCGAUUGCAGAAAAUGUUUCAAUAGAGAUGGCAAAACUUUCACGACGUGCCUCGUCGUCUGCAGCA ...-.........((((....))))...(((((..(((((((.((..(((.....))).)).))))))).............((((((.....))))))...))))) ( -19.60) >consensus U___UUUAA______AUAGAAAUCCCG__UUUUAUAUCUU_CUGCUUGCAGAAAAUGUCUUAAUAGAGAUGGCCAAACUUUCACGACGUGCCUCGUCGUCUGCAGCA .............................................(((((((....(((((....))))).............(((((.....)))))))))))).. (-16.89 = -16.62 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:19 2006