
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,399,507 – 4,399,749 |
| Length | 242 |
| Max. P | 0.940330 |

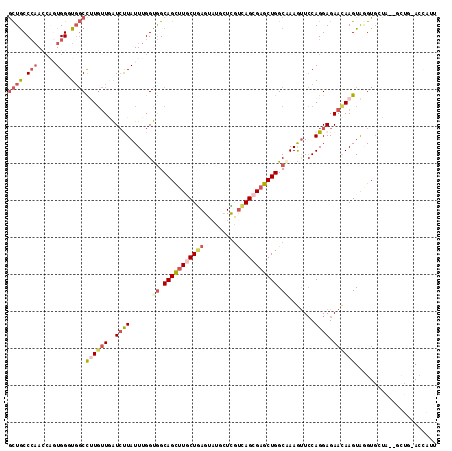
| Location | 4,399,507 – 4,399,620 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.60 |
| Mean single sequence MFE | -40.89 |
| Consensus MFE | -20.96 |
| Energy contribution | -23.02 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4399507 113 + 22407834 GCUGCCCAACCAGUGGUUGGCCUUGUUAAUCUUAUUUGGUGGCAGCUUGCUGAGUAUGCUCGUCAGCGAGCUGGCGAAAUUCCAGGAGAACAAGUAGGUGCUA--GCUG-ACCAAU ((((......))))(((..((...((....((((((((.((.((((((((((((......).))))))))))).))...(((.....))))))))))).))..--))..-)))... ( -40.60) >DroVir_CAF1 6271 116 + 1 AUUGCCACAGCUCUGGCUGCCUAUUCUCGUGCUAUUUGGCAGCAGCUUGCUGAGUCUCAUCAUCAGCGAGCUGGCCAAGCUGCAGGAGAACAAGUAAGUGCAGUUACCAAAUAUUU ((((((.((((....))))....(((((.(((..((((((..(((((((((((.........)))))))))))))))))..))).))))).......).)))))............ ( -46.90) >DroSim_CAF1 5165 113 + 1 GCUGCCCAACCAGUGGAUUGCCUUGUUGAUCUUAUUUGGUGGCAGCUUGCUGAGUAUGCUCGUCAGCGAGCUGGCAAAGUUCCAGGAGAACAAGUAUGUGCUA--GCUG-ACCAUU ((((((..(((((.(((((........)))))...))))))))))).....(((....)))((((((.(((..((...((((.....))))..))....))).--))))-)).... ( -39.40) >DroEre_CAF1 5170 113 + 1 GCUGCCCAACCAAUGGGUGGCCUUGUUGAUCUUAUUUGGUGGCAGCUUUCUGAGUAUGCUUGUUAGCGAGCUGGCAAAGUUCCAGGAGAACAAGUAGGUUACA--UUGG-ACCUUU ((..((((.....))))..))((((((..((((......((.((((((.((((.((....)))))).)))))).)).......)))).)))))).(((((...--...)-)))).. ( -36.82) >DroYak_CAF1 5171 113 + 1 GCUACCCAACCAGUGGGUGGCCUUGUUGAUCUUAUUUGGUGGCAGCUUGCUAAGUAUGCUUGUCAGCGAGCUGGCAAAGUUCCAGGAGAACAAGUAGGUGACA--GCUG-ACCAUA ((((((((.....))))))))...((((..(((((((..((.((((((((((((....)))...))))))))).))..((((.....)))))))))))...))--))..-...... ( -41.30) >DroAna_CAF1 7768 111 + 1 GCUGCCCAAAGUGUGGGUGGCCAUGUUAGUCCUAUUUGGCGGCAGUCUUCUGAGUAUGAUUGCCAGUGAGCUGGCCAAGUUCCAGGAGAAUAAGUGAGUGUUA--U-UC-ACAAU- ((..((((.....))))..))..((((..((((((((((((((((((..........)))))))((....)).)))))))...)))).)))).((((((...)--)-))-))...- ( -40.30) >consensus GCUGCCCAACCAGUGGGUGGCCUUGUUGAUCUUAUUUGGUGGCAGCUUGCUGAGUAUGCUCGUCAGCGAGCUGGCAAAGUUCCAGGAGAACAAGUAGGUGCUA__GCUG_ACCAUU ((((.(((.....))).))))((((((..((((......((.(((((((((((.........))))))))))).)).......)))).))))))...................... (-20.96 = -23.02 + 2.06)


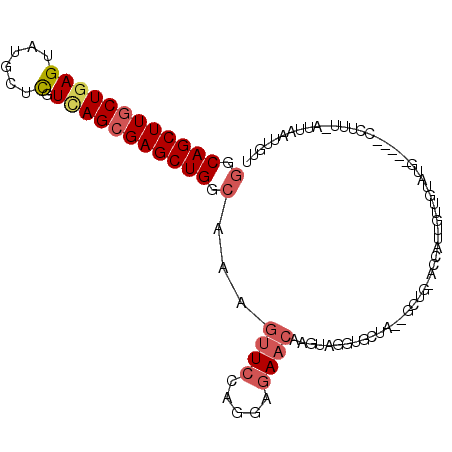
| Location | 4,399,547 – 4,399,643 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.10 |
| Mean single sequence MFE | -28.66 |
| Consensus MFE | -17.43 |
| Energy contribution | -18.10 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4399547 96 + 22407834 GGCAGCUUGCUGAGUAUGCUCGUCAGCGAGCUGGCGAAAUUCCAGGAGAACAAGUAGGUGCUA--GCUG-ACCAAUGUUCUAUG-----AUAAU-AUUAAAUGUU (.((((((((((((......).))))))))))).).......((..((((((.((.(((....--))).-))...)))))).))-----.....-.......... ( -28.80) >DroVir_CAF1 6311 104 + 1 AGCAGCUUGCUGAGUCUCAUCAUCAGCGAGCUGGCCAAGCUGCAGGAGAACAAGUAAGUGCAGUUACCAAAUAUUUGGUUGAGUUAUAU-AUUUAAUCAAUAUUU ..(((((((((((.........)))))))))))....(((((((..............)))))))...(((((((.((((((((.....-))))))))))))))) ( -32.84) >DroSec_CAF1 5209 96 + 1 GGCAGCUUGCUGAGUAUGCUCGUCAGCGAGCUGGCAAAGUUCCAGGAGAACAAGUAUGUGCUA--GCUG-ACCAUUGUUGUAUG-----CCUUU-AUUAAUUGUU (((((((.((.(((....))))).))).((((((((..((((.....)))).......)))))--))).-............))-----))...-.......... ( -27.10) >DroSim_CAF1 5205 96 + 1 GGCAGCUUGCUGAGUAUGCUCGUCAGCGAGCUGGCAAAGUUCCAGGAGAACAAGUAUGUGCUA--GCUG-ACCAUUGUUGUAUG-----CCUUU-AUUAAUUGUU (((((((.((.(((....))))).))).((((((((..((((.....)))).......)))))--))).-............))-----))...-.......... ( -27.10) >DroEre_CAF1 5210 96 + 1 GGCAGCUUUCUGAGUAUGCUUGUUAGCGAGCUGGCAAAGUUCCAGGAGAACAAGUAGGUUACA--UUGG-ACCUUUUUUGAGUU-----CCUCU-AUUAAGUGUA ((.((((((.(.(((.((((....)))).))).).))))))))(((((..((((.(((((...--...)-))))..))))..))-----)))..-.......... ( -26.70) >DroYak_CAF1 5211 96 + 1 GGCAGCUUGCUAAGUAUGCUUGUCAGCGAGCUGGCAAAGUUCCAGGAGAACAAGUAGGUGACA--GCUG-ACCAUAGUGGUAAU-----CCUCU-ACUAAUUGUA (((((((((((........(((((((....))))))).((((.....)))).))))(....))--))))-.)).((((((....-----...))-))))...... ( -29.40) >consensus GGCAGCUUGCUGAGUAUGCUCGUCAGCGAGCUGGCAAAGUUCCAGGAGAACAAGUAGGUGCUA__GCUG_ACCAUUGUUGUAUG_____CCUUU_AUUAAUUGUU (.((((((((((((......).))))))))))).)...((((.....))))...................................................... (-17.43 = -18.10 + 0.67)

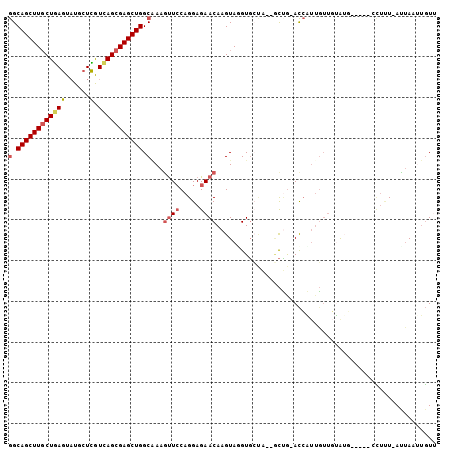
| Location | 4,399,643 – 4,399,749 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 74.83 |
| Mean single sequence MFE | -27.65 |
| Consensus MFE | -22.16 |
| Energy contribution | -22.22 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.30 |
| Mean z-score | -0.60 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.590940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4399643 106 + 22407834 --UGCUUUGUUUA--UUUCAGGGUUAACGCGCGCUAUCAGCGUCGGGCGCGUCUUGACUUCGGUACCAAAUUGGGCAUGAAUUCGCCCUUCUAAGAAUCACGUCAAAGAC --..(((((....--.(((.(((((((.(((((((..........))))))).)))))))............((((........))))......)))......))))).. ( -29.10) >DroVir_CAF1 6415 95 + 1 --UU-----U-GCUUUUGCAGAGUAAAUGCGCGCUAUCAGCGUCGGGCGCGACUUGAUUUUGGCACCAAGCUGGGCAUGAAUUCGCCAUUCUAAAUGUAACCA------- --..-----.-((((.((((((((.....((((((..........)))))).....))))).)))..))))..(((........)))................------- ( -22.70) >DroEre_CAF1 5306 106 + 1 --UUCCUUGUUUA--UUUUAGGGUUAACGCGCGCUAUCAGCGUCGUGCGCGUCUUGACUUUGGUACCAAAUUGGGCAUGAAUUCGCCCUUCUAAGAAUCACGGCUAAGAC --..(((((....--...))))).....((((((..........))))))((((((.((.((((........((((........))))........)))).)).)))))) ( -28.99) >DroYak_CAF1 5307 106 + 1 --UUCCACGUUUA--UUUUAGGGUUAACGCGCGCUACCAGCGUCGAGCGCGUCUUGACUUUGGUACCAAGUUGGGCAUGAAUUCGCCCUUCUAAGAAUCACGGCUAUGAC --.....(((...--((((((((((((.(((((((..........))))))).)))))((((....))))..((((........)))).)))))))...)))........ ( -29.60) >DroMoj_CAF1 5601 93 + 1 ---------U-UCCUUUGCAGAGUUAAUGCGCGCUAUCAGCGUCGAGCGCGUCUUGAUUUUGGCACCAAGCUGGGCAUGAAUUCGCCAUUUUAAAAGCUACCU------- ---------.-.....(((((((((((.(((((((..........))))))).)))))))).)))...((((.(((........)))........))))....------- ( -26.50) >DroAna_CAF1 7901 99 + 1 UCUAUUCUAUCUA--UUUUAGGGUCAAUGCGCGCUAUCAGCGGCGGGCACGCCUUGAUUUUGGCACCAAACUGGGCAUGAACUCGCCCUUCUAAGAAU---------GAU ........(((..--(((((((((...(((.((((......)))).))).(((........)))))).....((((........))))..))))))..---------))) ( -29.00) >consensus __UUC__UGUUUA__UUUCAGGGUUAACGCGCGCUAUCAGCGUCGGGCGCGUCUUGACUUUGGCACCAAACUGGGCAUGAAUUCGCCCUUCUAAGAAUCACGG____GAC ..................(((((((((.(((((((..........))))))).)))))))))..........((((........))))...................... (-22.16 = -22.22 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:16 2006