
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,393,637 – 4,393,735 |
| Length | 98 |
| Max. P | 0.688128 |

| Location | 4,393,637 – 4,393,735 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 112 |
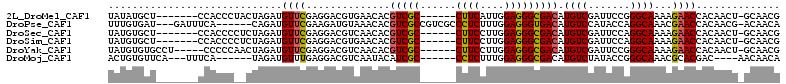
| Reading direction | reverse |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -31.85 |
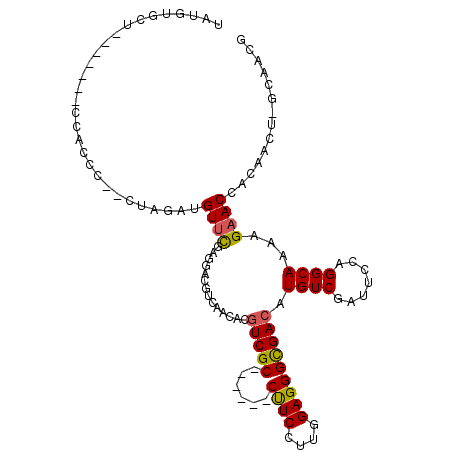
| Consensus MFE | -15.27 |
| Energy contribution | -15.10 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4393637 98 - 22407834 UAUAUGCU-------CCACCCUACUAGAUGUUCGAGGACGUGAACACGUCGC------CUUCAUUGGAGGGCGACAUGUCGAUUCCGGGCAAAAGAACCACAACU-GCAACG ....(((.-------.............((((((.(((((...(((.(((((------((((....))))))))).))))).)))))))))..............-)))... ( -28.69) >DroPse_CAF1 6520 102 - 1 UUUGUGAU---GAUUUCA------CAGAUGUUCGAAGAUGUAAACACGUCGCCGUCGCCCUCUUUGGAGGGUGACAUGUCCAUACCAGGCAAACGAACCACAACG-ACAACA (((((((.---....)))------)))).(((((..(((((....)))))((((((((((((....))))))))).((.......)))))...))))).......-...... ( -37.60) >DroSec_CAF1 4849 98 - 1 UAUGUGCU-------CCACCCCUCUAGAUGUUCGAGGACGUCAACACGUCGC------CUUCCUUGGAGGGCGACAUGUCGAUUCCAGGCAAAAGAACCACAACU-GCAACG ..((((..-------...........((((((....)))))).....(((((------((((....))))))))).((((.......)))).......))))...-...... ( -30.50) >DroSim_CAF1 4911 98 - 1 UAUGUGCU-------CCACCCCUCUAGAUGUUCGAGGACGUGAACACGUCGC------CUUCCUUGGAGGGCGACAUGUCGAUUCCAGGCAAAAGAACCACAACU-GCAACG ....(((.-------.....((((.........))))..(((.....(((((------((((....))))))))).((((.......)))).......)))....-)))... ( -31.00) >DroYak_CAF1 4901 100 - 1 UAUGUGUGCCU-----CCCCCAACUAGAUGUUCGAGGACGUCAACACGUCGC------CUUCCUUGGAGGGCGACAUGUCGAUUCCGGGCAAAAGAACCACAACU-GCAACG ..(((((((((-----...(.(((.....))).).(((.(((.(((.(((((------((((....))))))))).))).))))))))))).......))))...-...... ( -34.31) >DroMoj_CAF1 5660 93 - 1 ACUGUGUUCA---UUUCA------UAGAUGUUUGAGGACGUCAAUACAUCGC------CCUCUUUGGAGGGCGACAUGUCUAUACCGGGCAAACGCACGAC----AACAACA ..((((((..---.....------..((((((....))))))......((((------((((....))))))))..(((((.....)))))))))))....----....... ( -29.00) >consensus UAUGUGCU_______CCACCC__CUAGAUGUUCGAGGACGUCAACACGUCGC______CUUCCUUGGAGGGCGACAUGUCGAUUCCAGGCAAAAGAACCACAACU_GCAACG .............................((((..............(((((......((((....))))))))).((((.......))))...)))).............. (-15.27 = -15.10 + -0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:14 2006