| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,372,564 – 4,372,664 |
| Length | 100 |
| Max. P | 0.998755 |

| Location | 4,372,564 – 4,372,664 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 83.50 |
| Mean single sequence MFE | -33.19 |
| Consensus MFE | -26.38 |
| Energy contribution | -27.70 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.82 |
| Structure conservation index | 0.79 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
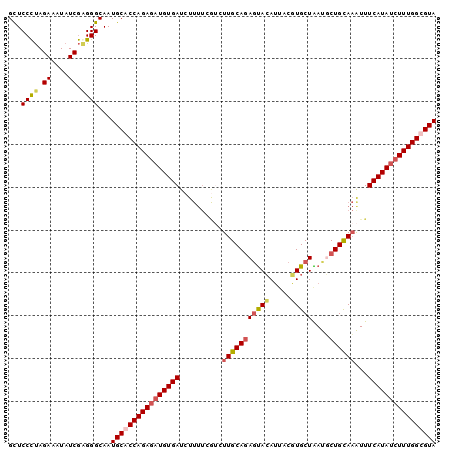
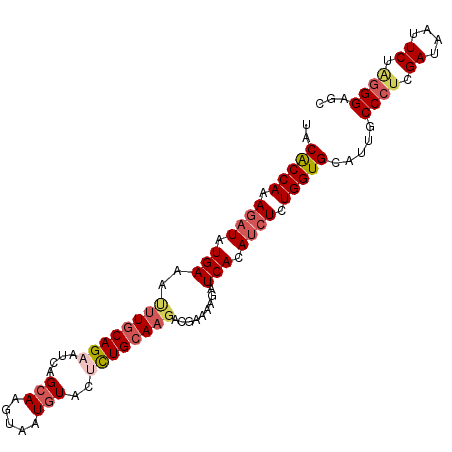
>2L_DroMel_CAF1 4372564 100 + 22407834 GCUCCCUAGAAAUAUCGAGGGCAAUGCACCAGAGAUGUGAUCUUUUCGUCUUGCAGAUUACAUUACGUGCUAAUGCUGCAAAUUUCAUAUCUUUGGUGUA ...((((.((....)).))))...((((((((((((((((..........(((((((((((((...))).)))).))))))...)))))))))))))))) ( -34.32) >DroSec_CAF1 24438 92 + 1 GCUCCCUAGAAAUAUCGAGGGCAAUGCACCAGAUAUGUGAUC--------UUGCAAAGUACAUAACUUGCUAAUGCUGUACAUUUCAUAUCUUUGGUGUA ...((((.((....)).))))...(((((((((.((((((..--------.((((((((.....))))((....))))))....)))))).))))))))) ( -26.00) >DroEre_CAF1 19762 100 + 1 GUUCCUCAGAAUUAUCAAGGGCAAUGCACCAGAGAUGUGAUCUUUUCGUCUUGCAGAGCACAUUACGUGCUGAUUCUGCAAAUUUCAUAUCUUUGGCGUA ((((((..((....))..))).)))((.((((((((((((..........(((((((((((.....))))....)))))))...)))))))))))).)). ( -31.12) >DroYak_CAF1 20140 100 + 1 GCUCCCUAGAAUUAUCGGGGGCAAUGCACCAGAGCUGUGAUCUUUUCAGCUUGCAGAGUAGAAUACUUGCUGACUCUGCAAGUUUCAUAUCUUUGGCGUA ((((((..........))))))..(((.((((((.(((((.......((((((((((((((........)).))))))))))))))))).)))))).))) ( -41.31) >consensus GCUCCCUAGAAAUAUCGAGGGCAAUGCACCAGAGAUGUGAUCUUUUCGUCUUGCAGAGUACAUUACGUGCUAAUGCUGCAAAUUUCAUAUCUUUGGCGUA ...((((.((....)).))))...((((((((((((((((..........(((((((((((.....)))))....))))))...)))))))))))))))) (-26.38 = -27.70 + 1.31)


| Location | 4,372,564 – 4,372,664 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 83.50 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -15.81 |
| Energy contribution | -17.62 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
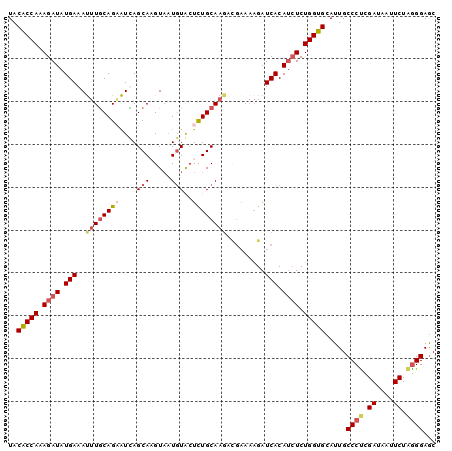
>2L_DroMel_CAF1 4372564 100 - 22407834 UACACCAAAGAUAUGAAAUUUGCAGCAUUAGCACGUAAUGUAAUCUGCAAGACGAAAAGAUCACAUCUCUGGUGCAUUGCCCUCGAUAUUUCUAGGGAGC ..(((((.((((.(((..(((((((.((((.((.....))))))))))))).(.....).))).)))).))))).....((((.((....)).))))... ( -27.00) >DroSec_CAF1 24438 92 - 1 UACACCAAAGAUAUGAAAUGUACAGCAUUAGCAAGUUAUGUACUUUGCAA--------GAUCACAUAUCUGGUGCAUUGCCCUCGAUAUUUCUAGGGAGC ..(((((..((((((.((((.....)))).((((((.....)).))))..--------.....))))))))))).....((((.((....)).))))... ( -24.30) >DroEre_CAF1 19762 100 - 1 UACGCCAAAGAUAUGAAAUUUGCAGAAUCAGCACGUAAUGUGCUCUGCAAGACGAAAAGAUCACAUCUCUGGUGCAUUGCCCUUGAUAAUUCUGAGGAAC ..(((((.((((.(((..((((((((....((((.....)))))))))))).(.....).))).)))).)))))......((((((....)).))))... ( -28.00) >DroYak_CAF1 20140 100 - 1 UACGCCAAAGAUAUGAAACUUGCAGAGUCAGCAAGUAUUCUACUCUGCAAGCUGAAAAGAUCACAGCUCUGGUGCAUUGCCCCCGAUAAUUCUAGGGAGC ..(((((.((...(((..((((((((((.((........))))))))))))((....)).)))...)).)))))....((.(((..........))).)) ( -30.40) >consensus UACACCAAAGAUAUGAAAUUUGCAGAAUCAGCAAGUAAUGUACUCUGCAAGACGAAAAGAUCACAUCUCUGGUGCAUUGCCCUCGAUAAUUCUAGGGAGC ..(((((.((((.(((..((((((((....(((.....)))..)))))))).........))).)))).))))).....((((.((....)).))))... (-15.81 = -17.62 + 1.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:11 2006