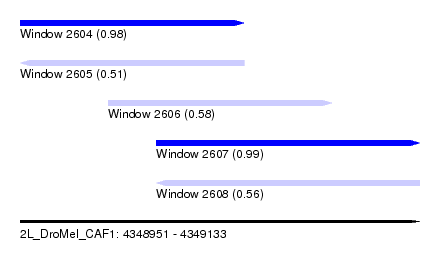
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,348,951 – 4,349,133 |
| Length | 182 |
| Max. P | 0.993826 |

| Location | 4,348,951 – 4,349,053 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.58 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -32.20 |
| Energy contribution | -33.82 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4348951 102 + 22407834 UCUGUUAACUACGCCGCCAUUUGUCUGCAGUUGGUAAGAUGGUAGCUUAGUCACUUGGC------------------AACUGGCAACAACUAACUGUUAACUGGGCCCAGACCUGGGCCA ...((((((......((((..((.(((.(((((.(.....).)))))))).))..))))------------------...((....)).......))))))..((((((....)))))). ( -34.10) >DroSec_CAF1 17662 112 + 1 UCUGUUAACUUUGCCGCCAUUUGUCUGCAGUUGGUAAGAUGGUAGCUUAGUCACUUGGCAGUGCG---GCAACUGCCAACUGGCAACAACUAACUGUUAACUGGG-----ACCUCGGCCA ............((((((....((..((((((((((((.......))).((((.((((((((...---...)))))))).))))....)))))))))..))..))-----....)))).. ( -37.30) >DroSim_CAF1 18409 117 + 1 UCUGUUAACUUUGCCGCCAUUUGUCUGCAGUUGGUAAGAUGGUAGCUUAGUCACUUGGCAGUGCG---GCAACUGCCAACUGGCAACAACUAACUGUUAACUGGGCCCAGACCUGGGCCA ...((((((...((..((((((..((......))..))))))..))...((((.((((((((...---...)))))))).))))...........))))))..((((((....)))))). ( -46.80) >DroEre_CAF1 22916 117 + 1 UCUGUUAACUUUGCCGACAGUUGUCUGCAGUUGGUAAGAUGGUGGCUUAGUCACUUGGCAGCUCG---GCAACUGCCAACUGGCAACAACUAACUGUUAACUGGGCCUAGACCUGGGCCA ...((((((...((.(((....))).))(((((((.....((((((...))))))((((((....---....))))))..((....)))))))))))))))..((((((....)))))). ( -43.00) >DroYak_CAF1 19164 120 + 1 UCUGUUAACUUUGCCGCCAUUUGUCUGCAGUUGGUAUGAUGGUAGCUUAGUCACUUGGCAGCUCGGCGGCAACUGCCAACUGGCAACAACUAACUGUUAACUGGGCUUAGACCUGGGCCA ...((((((.((((((((..(..(((((.....))).))..).((((..(((....))))))).)))))))).((((....))))..........))))))..((((((....)))))). ( -41.80) >consensus UCUGUUAACUUUGCCGCCAUUUGUCUGCAGUUGGUAAGAUGGUAGCUUAGUCACUUGGCAGCGCG___GCAACUGCCAACUGGCAACAACUAACUGUUAACUGGGCCCAGACCUGGGCCA ...((((((...((.((((((...((......))...)))))).))...((((.(((((((...........))))))).))))...........))))))..((((((....)))))). (-32.20 = -33.82 + 1.62)

| Location | 4,348,951 – 4,349,053 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.58 |
| Mean single sequence MFE | -36.14 |
| Consensus MFE | -26.68 |
| Energy contribution | -28.11 |
| Covariance contribution | 1.43 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.510307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4348951 102 - 22407834 UGGCCCAGGUCUGGGCCCAGUUAACAGUUAGUUGUUGCCAGUU------------------GCCAAGUGACUAAGCUACCAUCUUACCAACUGCAGACAAAUGGCGGCGUAGUUAACAGA .((((((....))))))..((((((.......((((((((.((------------------((..(((...((((.......))))...))))))).....))))))))..))))))... ( -31.40) >DroSec_CAF1 17662 112 - 1 UGGCCGAGGU-----CCCAGUUAACAGUUAGUUGUUGCCAGUUGGCAGUUGC---CGCACUGCCAAGUGACUAAGCUACCAUCUUACCAACUGCAGACAAAUGGCGGCAAAGUUAACAGA .(((....))-----)...((((((......((((((((((((.(((((((.---..((((....))))..((((.......)))).))))))).)))...))))))))).))))))... ( -33.50) >DroSim_CAF1 18409 117 - 1 UGGCCCAGGUCUGGGCCCAGUUAACAGUUAGUUGUUGCCAGUUGGCAGUUGC---CGCACUGCCAAGUGACUAAGCUACCAUCUUACCAACUGCAGACAAAUGGCGGCAAAGUUAACAGA .((((((....))))))..((((((......((((((((((((.(((((((.---..((((....))))..((((.......)))).))))))).)))...))))))))).))))))... ( -43.20) >DroEre_CAF1 22916 117 - 1 UGGCCCAGGUCUAGGCCCAGUUAACAGUUAGUUGUUGCCAGUUGGCAGUUGC---CGAGCUGCCAAGUGACUAAGCCACCAUCUUACCAACUGCAGACAACUGUCGGCAAAGUUAACAGA .((((........))))..((((((....(((((....((.(((((((((..---..))))))))).))..((((.......)))).)))))((.(((....))).))...))))))... ( -37.40) >DroYak_CAF1 19164 120 - 1 UGGCCCAGGUCUAAGCCCAGUUAACAGUUAGUUGUUGCCAGUUGGCAGUUGCCGCCGAGCUGCCAAGUGACUAAGCUACCAUCAUACCAACUGCAGACAAAUGGCGGCAAAGUUAACAGA .(((....)))........((((((.........(((((....)))))(((((((((..((((..(((.....................))))))).....))))))))).))))))... ( -35.20) >consensus UGGCCCAGGUCUAGGCCCAGUUAACAGUUAGUUGUUGCCAGUUGGCAGUUGC___CGAACUGCCAAGUGACUAAGCUACCAUCUUACCAACUGCAGACAAAUGGCGGCAAAGUUAACAGA .((((........))))..((((((..(((((......((.((((((((.........)))))))).)))))))(((.((((..................)))).)))...))))))... (-26.68 = -28.11 + 1.43)



| Location | 4,348,991 – 4,349,093 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.91 |
| Mean single sequence MFE | -36.94 |
| Consensus MFE | -29.04 |
| Energy contribution | -30.26 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4348991 102 + 22407834 GGUAGCUUAGUCACUUGGC------------------AACUGGCAACAACUAACUGUUAACUGGGCCCAGACCUGGGCCAGGUACGCCCCCAACAUGCAUUCCAACAUGCCAUGCGAAAG ((..(((((((...(((.(------------------....).)))..)))))......(((.((((((....)))))).)))..))..))..(((((((......))).))))...... ( -27.90) >DroSec_CAF1 17702 112 + 1 GGUAGCUUAGUCACUUGGCAGUGCG---GCAACUGCCAACUGGCAACAACUAACUGUUAACUGGG-----ACCUCGGCCAGGUACGCCCCCAACAUGCAUUCCGACAUGCCAUGCGAAAG ((..((...((((.((((((((...---...)))))))).))))...........((...((((.-----.......))))..))))..))..(((((((......))).))))...... ( -33.40) >DroSim_CAF1 18449 117 + 1 GGUAGCUUAGUCACUUGGCAGUGCG---GCAACUGCCAACUGGCAACAACUAACUGUUAACUGGGCCCAGACCUGGGCCAGGUACGCCCCCAACAUGCAUUCCAACAUGCCAUGCGAAAG ((..((...((((.((((((((...---...)))))))).))))((((......)))).(((.((((((....)))))).)))..))..))..(((((((......))).))))...... ( -42.50) >DroEre_CAF1 22956 116 + 1 GGUGGCUUAGUCACUUGGCAGCUCG---GCAACUGCCAACUGGCAACAACUAACUGUUAACUGGGCCUAGACCUGGGCCAGGC-CGCCCCCAACAUGCAUUCCAACAUGCCAUGCGAAAG ((((((((.((((.(((((((....---....))))))).))))((((......)))).....((((((....))))))))))-)))).....(((((((......))).))))...... ( -43.50) >DroYak_CAF1 19204 119 + 1 GGUAGCUUAGUCACUUGGCAGCUCGGCGGCAACUGCCAACUGGCAACAACUAACUGUUAACUGGGCUUAGACCUGGGCCAGGC-CGCCCCCAACAUGCAUUCCAACAUGCCAUGCGAAAG ((..((...(((....))).....((((((....)))..(((((((((......))))..(..((......))..))))))))-)))..))..(((((((......))).))))...... ( -37.40) >consensus GGUAGCUUAGUCACUUGGCAGCGCG___GCAACUGCCAACUGGCAACAACUAACUGUUAACUGGGCCCAGACCUGGGCCAGGUACGCCCCCAACAUGCAUUCCAACAUGCCAUGCGAAAG ((..((...((((.(((((((...........))))))).))))((((......))))..((((.((((....))))))))....))..))..(((((((......))).))))...... (-29.04 = -30.26 + 1.22)


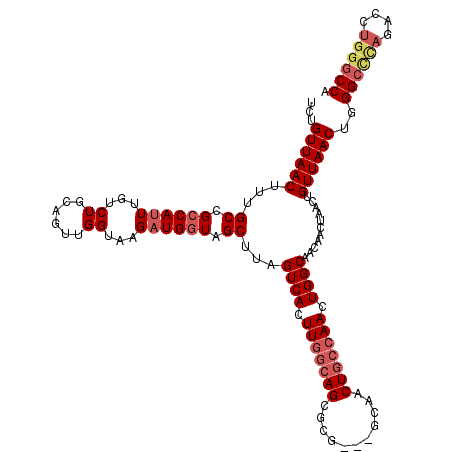
| Location | 4,349,013 – 4,349,133 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.32 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -27.96 |
| Energy contribution | -28.16 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4349013 120 + 22407834 UGGCAACAACUAACUGUUAACUGGGCCCAGACCUGGGCCAGGUACGCCCCCAACAUGCAUUCCAACAUGCCAUGCGAAAGGCAAAUCUAACUGCAUGUUGCAUGCCACUCCAAGACAAAA (((((..............(((.((((((....)))))).))).......(((((((((........((((........))))........)))))))))..)))))............. ( -38.59) >DroSec_CAF1 17739 115 + 1 UGGCAACAACUAACUGUUAACUGGG-----ACCUCGGCCAGGUACGCCCCCAACAUGCAUUCCGACAUGCCAUGCGAAAGUCAAACCUAACUGCAUGUUGCAUGCCACUCCAAGACAAAA (((((..........((...((((.-----.......))))..)).....(((((((((........((....((....))))........)))))))))..)))))............. ( -25.29) >DroSim_CAF1 18486 120 + 1 UGGCAACAACUAACUGUUAACUGGGCCCAGACCUGGGCCAGGUACGCCCCCAACAUGCAUUCCAACAUGCCAUGCGAAAGUCAAAUCUAACUGCAUGUUGCAUGCCACUGCAAGACAAAA .(((((((......)))).(((.((((((....)))))).)))..))).......((((......(((((((((((..((......))...))))))..)))))....))))........ ( -34.90) >DroEre_CAF1 22993 118 + 1 UGGCAACAACUAACUGUUAACUGGGCCUAGACCUGGGCCAGGC-CGCCCCCAACAUGCAUUCCAACAUGCCAUGCGAAAGUCAAAUCUAACUGCAUGUUGCAUGCCACUCCAAGACAA-A .(((((((......))))..((((.((((....))))))))))-)((...(((((((((........((....((....))))........)))))))))...)).............-. ( -31.39) >DroYak_CAF1 19244 107 + 1 UGGCAACAACUAACUGUUAACUGGGCUUAGACCUGGGCCAGGC-CGCCCCCAACAUGCAUUCCAACAUGCCAUGCGAAAGUCGAAUCUAACUGCAUGUUGCAUGCCGC------------ .(((((((......))))..((((.((.......)).))))))-)...........((((..((((((((...((....))...........)))))))).))))...------------ ( -28.84) >consensus UGGCAACAACUAACUGUUAACUGGGCCCAGACCUGGGCCAGGUACGCCCCCAACAUGCAUUCCAACAUGCCAUGCGAAAGUCAAAUCUAACUGCAUGUUGCAUGCCACUCCAAGACAAAA .(((((((......))))..((((.((((....))))))))....)))........((((..((((((((..(((....).)).........)))))))).))))............... (-27.96 = -28.16 + 0.20)


| Location | 4,349,013 – 4,349,133 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.32 |
| Mean single sequence MFE | -39.66 |
| Consensus MFE | -33.82 |
| Energy contribution | -33.98 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4349013 120 - 22407834 UUUUGUCUUGGAGUGGCAUGCAACAUGCAGUUAGAUUUGCCUUUCGCAUGGCAUGUUGGAAUGCAUGUUGGGGGCGUACCUGGCCCAGGUCUGGGCCCAGUUAACAGUUAGUUGUUGCCA .............(((((.(((((..((.(((((....(((((..(((((.(((......))))))))..)))))....((((((((....)))).))))))))).))..)))))))))) ( -44.10) >DroSec_CAF1 17739 115 - 1 UUUUGUCUUGGAGUGGCAUGCAACAUGCAGUUAGGUUUGACUUUCGCAUGGCAUGUCGGAAUGCAUGUUGGGGGCGUACCUGGCCGAGGU-----CCCAGUUAACAGUUAGUUGUUGCCA .............(((((.(((((..((.(((((.(..((((((.((..(((((((......))))))).(((.....))).)).)))))-----)..).))))).))..)))))))))) ( -34.20) >DroSim_CAF1 18486 120 - 1 UUUUGUCUUGCAGUGGCAUGCAACAUGCAGUUAGAUUUGACUUUCGCAUGGCAUGUUGGAAUGCAUGUUGGGGGCGUACCUGGCCCAGGUCUGGGCCCAGUUAACAGUUAGUUGUUGCCA .....(((.(((...((((.((((((((.....((........)).....))))))))..)))).))).)))(((....((((((((....)))).))))..(((((....)))))))). ( -41.10) >DroEre_CAF1 22993 118 - 1 U-UUGUCUUGGAGUGGCAUGCAACAUGCAGUUAGAUUUGACUUUCGCAUGGCAUGUUGGAAUGCAUGUUGGGGGCG-GCCUGGCCCAGGUCUAGGCCCAGUUAACAGUUAGUUGUUGCCA .-....((..(....((((.((((((((.....((........)).....))))))))..))))...)..))((((-(((((((....).))))))).....(((((....)))))))). ( -41.40) >DroYak_CAF1 19244 107 - 1 ------------GCGGCAUGCAACAUGCAGUUAGAUUCGACUUUCGCAUGGCAUGUUGGAAUGCAUGUUGGGGGCG-GCCUGGCCCAGGUCUAAGCCCAGUUAACAGUUAGUUGUUGCCA ------------..((((.(((((((((((((......))))...)))).((((......)))).(((((.(((((-(((((...))))))...))))...)))))....))))))))). ( -37.50) >consensus UUUUGUCUUGGAGUGGCAUGCAACAUGCAGUUAGAUUUGACUUUCGCAUGGCAUGUUGGAAUGCAUGUUGGGGGCGUACCUGGCCCAGGUCUAGGCCCAGUUAACAGUUAGUUGUUGCCA ..............((((.(((((((((.(..((......))..))))).((((......)))).(((((.((((..(((((...)))))....))))...)))))....))))))))). (-33.82 = -33.98 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:58 2006