
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,347,006 – 4,347,165 |
| Length | 159 |
| Max. P | 0.974951 |

| Location | 4,347,006 – 4,347,125 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -38.46 |
| Consensus MFE | -33.58 |
| Energy contribution | -33.70 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4347006 119 + 22407834 AGUGGAAGAGCCAUUGAGUCGGAGAGGGAUGGAGAG-AUGUGUGGAGAAUCAUCAAAGGUUUGCCGCAGCGGCUUUUCCGCCUUUGUGGCAAAUCCUCAAAGGAAUUUUCCGAUAUUUGU (((((.....)))))..(((((((((.....(((.(-(((..........))))...((((((((((((.(((......))).))))))))))))))).......)))))))))...... ( -42.50) >DroSec_CAF1 15720 119 + 1 AGAGGAAGAGCCAGCGAGUCAGAGAGGGAUGGAGAG-AUGUGUGGAGAAUCAUCAAAGGUUUGCCGCAGCGGCUUUUCCGCCUUUGUGGCAAAUCCUCAAAGGAAUUUUCCGAUAUUUGU .(((((...(....)..((((.(((((..(((((((-.(((((((..((((......))))..))))).))..)))))))))))).))))...)))))...(((....)))......... ( -38.30) >DroSim_CAF1 16480 119 + 1 AGAGGAAGAGCCAUCGAGUCAGAGAGGGAUGGAGAG-AUGUGUGGAGAAUCAUCAAAGGUUUGCCGCAGCGGCUUUUCCGCCUUUGUGGCAAAUCCUCAAAGGAAUUUUCCGAUAUUUGU .(((((.((....))..((((.(((((..(((((((-.(((((((..((((......))))..))))).))..)))))))))))).))))...)))))...(((....)))......... ( -39.40) >DroEre_CAF1 16080 120 + 1 AGAGGAACAGACAUUGAGUCGGAGAGGGAUGGAGAGCGUGUGUGUGGAAUCAUCAAAGGUUUGCCGCAGCGGCUUUUCCGUCUUUGUGGCAAAUCCUCAAAGGAAUUUUCCGAUAUUUGU .(((((...(((.....)))..(.(((((((((((((.(((.(((((.(((......)))...)))))))))).))))))))))).)......)))))...(((....)))......... ( -36.50) >DroYak_CAF1 16393 119 + 1 AGGGGAAGAGACAGCGAGUCGGAGAGGGAUGGCGAG-AUGUGUGGAGAAUCAUCAAAGGUUUGCCGCAGCGGCUUUUCCGUCUUUGUGGCAAAUCCUCAAAGGAAUUUUCCGAUAUUUGU .............((((((((((((((((.(((..(-...(((((..((((......))))..))))).).))).)))).((((((.((.....)).))))))...))))))))..)))) ( -35.60) >consensus AGAGGAAGAGCCAUCGAGUCGGAGAGGGAUGGAGAG_AUGUGUGGAGAAUCAUCAAAGGUUUGCCGCAGCGGCUUUUCCGCCUUUGUGGCAAAUCCUCAAAGGAAUUUUCCGAUAUUUGU ...((((((........((((.(((((..((((((...(((((((.(((((......))))).))))).))...))))))))))).))))...(((.....))).))))))......... (-33.58 = -33.70 + 0.12)



| Location | 4,347,006 – 4,347,125 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -24.19 |
| Energy contribution | -24.35 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4347006 119 - 22407834 ACAAAUAUCGGAAAAUUCCUUUGAGGAUUUGCCACAAAGGCGGAAAAGCCGCUGCGGCAAACCUUUGAUGAUUCUCCACACAU-CUCUCCAUCCCUCUCCGACUCAAUGGCUCUUCCACU .........((((....((.(((((..((((((.((..(((......)))..)).)))))).....((((..........)))-).................))))).))...))))... ( -29.50) >DroSec_CAF1 15720 119 - 1 ACAAAUAUCGGAAAAUUCCUUUGAGGAUUUGCCACAAAGGCGGAAAAGCCGCUGCGGCAAACCUUUGAUGAUUCUCCACACAU-CUCUCCAUCCCUCUCUGACUCGCUGGCUCUUCCUCU .........(((....)))...(((((...((((....(((((.....)))))((((((.......((((.............-.....))))......)).).)))))))...))))). ( -27.89) >DroSim_CAF1 16480 119 - 1 ACAAAUAUCGGAAAAUUCCUUUGAGGAUUUGCCACAAAGGCGGAAAAGCCGCUGCGGCAAACCUUUGAUGAUUCUCCACACAU-CUCUCCAUCCCUCUCUGACUCGAUGGCUCUUCCUCU .........(((.((((.....((((.((((((.((..(((......)))..)).))))))))))....)))).)))......-....(((((............))))).......... ( -30.10) >DroEre_CAF1 16080 120 - 1 ACAAAUAUCGGAAAAUUCCUUUGAGGAUUUGCCACAAAGACGGAAAAGCCGCUGCGGCAAACCUUUGAUGAUUCCACACACACGCUCUCCAUCCCUCUCCGACUCAAUGUCUGUUCCUCU .........((((.........((((.((((((....((.(((.....)))))..)))))).....((((...................))))))))...(((.....)))..))))... ( -24.41) >DroYak_CAF1 16393 119 - 1 ACAAAUAUCGGAAAAUUCCUUUGAGGAUUUGCCACAAAGACGGAAAAGCCGCUGCGGCAAACCUUUGAUGAUUCUCCACACAU-CUCGCCAUCCCUCUCCGACUCGCUGUCUCUUCCCCU .........((((.........((((.((((((....((.(((.....)))))..)))))).....((((..........)))-)........))))...(((.....)))..))))... ( -24.60) >consensus ACAAAUAUCGGAAAAUUCCUUUGAGGAUUUGCCACAAAGGCGGAAAAGCCGCUGCGGCAAACCUUUGAUGAUUCUCCACACAU_CUCUCCAUCCCUCUCCGACUCGAUGGCUCUUCCUCU .......(((((..........((((.((((((.((..(((......)))..)).)))))))))).((((...................))))....))))).................. (-24.19 = -24.35 + 0.16)

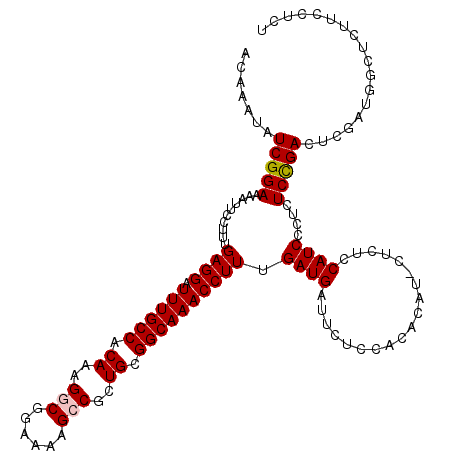
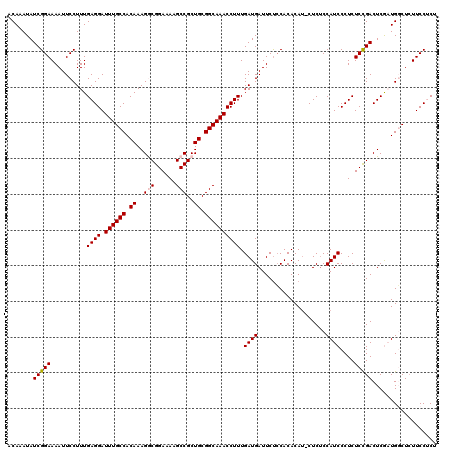
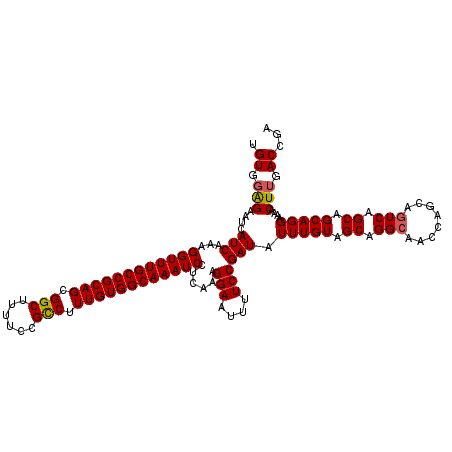
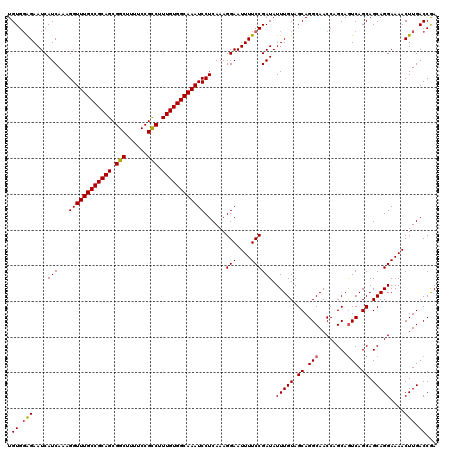
| Location | 4,347,045 – 4,347,165 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -36.24 |
| Consensus MFE | -35.04 |
| Energy contribution | -35.04 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4347045 120 + 22407834 UGUGGAGAAUCAUCAAAGGUUUGCCGCAGCGGCUUUUCCGCCUUUGUGGCAAAUCCUCAAAGGAAUUUUCCGAUAUUUGUAGCAGGCAACCAGCAAUCAGCAGCAGGAAAACUUGACCGA ..(((((((((.....(((((((((((((.(((......))).)))))))))).))).....).))))))))...(((((.((..((.....)).....)).)))))............. ( -35.90) >DroSec_CAF1 15759 120 + 1 UGUGGAGAAUCAUCAAAGGUUUGCCGCAGCGGCUUUUCCGCCUUUGUGGCAAAUCCUCAAAGGAAUUUUCCGAUAUUUGUAGCAGGCAACCAGCAGUCAGCAGCAGGAAAACUUGACCGA ..(((((((((.....(((((((((((((.(((......))).)))))))))).))).....).))))))))...(((((.((.(((........))).)).)))))............. ( -38.00) >DroSim_CAF1 16519 120 + 1 UGUGGAGAAUCAUCAAAGGUUUGCCGCAGCGGCUUUUCCGCCUUUGUGGCAAAUCCUCAAAGGAAUUUUCCGAUAUUUGUAGCAGGCAACCAGCAGUCAGCAGCAGGAAAACUUGACCGA ..(((((((((.....(((((((((((((.(((......))).)))))))))).))).....).))))))))...(((((.((.(((........))).)).)))))............. ( -38.00) >DroEre_CAF1 16120 120 + 1 UGUGUGGAAUCAUCAAAGGUUUGCCGCAGCGGCUUUUCCGUCUUUGUGGCAAAUCCUCAAAGGAAUUUUCCGAUAUUUGUAGCAGGCAACCAGCAGUCAGCAGCAGGAAAACUUGACCAA ....(((....(((...((((((((((((.(((......))).))))))))))))......(((....)))))).(((((.((.(((........))).)).))))).........))). ( -34.00) >DroYak_CAF1 16432 120 + 1 UGUGGAGAAUCAUCAAAGGUUUGCCGCAGCGGCUUUUCCGUCUUUGUGGCAAAUCCUCAAAGGAAUUUUCCGAUAUUUGUAGCAGGCAACCAGCAGUCAGCAGCAGGAAAACUUGACCAA ..(((((((((.....(((((((((((((.(((......))).)))))))))).))).....).))))))))...(((((.((.(((........))).)).)))))............. ( -35.30) >consensus UGUGGAGAAUCAUCAAAGGUUUGCCGCAGCGGCUUUUCCGCCUUUGUGGCAAAUCCUCAAAGGAAUUUUCCGAUAUUUGUAGCAGGCAACCAGCAGUCAGCAGCAGGAAAACUUGACCGA .((.(((....(((...((((((((((((.(((......))).))))))))))))......(((....)))))).(((((.((.(((........))).)).)))))....))).))... (-35.04 = -35.04 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:51 2006