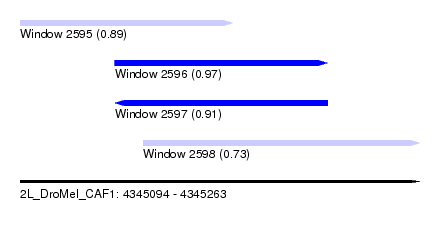
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,345,094 – 4,345,263 |
| Length | 169 |
| Max. P | 0.972402 |

| Location | 4,345,094 – 4,345,184 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 93.96 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -22.10 |
| Energy contribution | -21.98 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4345094 90 + 22407834 CAGCAGCGUGCAAUCAAACGGAAGUUAGGGCGAGCGUGUAAAGCGGAAACGGAAAC--AGAAAUCAUUCCCUUGUGAUAGCAAGCGGUGCAG ..(((.(((((.(((((((....)))((((...((.......))(....)(....)--..........))))..)))).))..))).))).. ( -26.20) >DroSec_CAF1 13835 90 + 1 ACGCAGCGUGCAAUCAAACGGAAGUUAGGGCGAGCGUGGAAAGCGGAAACGGAAAC--AGAAAUCAUUCCCUUGUGAUAGAAAGCGGUGCAG ..(((.(((.(.(((.(((....)))...(((((...((((...(....)(....)--........)))))))))))).)...))).))).. ( -25.80) >DroSim_CAF1 14588 90 + 1 ACGCAGCGUGCAAUCAAACGGAAGUUAGGGCGAGCGUGGAAAGCGGAAACGGAAAC--AGAAAUCAUUCCCUUGUGAUAGAAAGCGGUGCAG ..(((.(((.(.(((.(((....)))...(((((...((((...(....)(....)--........)))))))))))).)...))).))).. ( -25.80) >DroYak_CAF1 14398 92 + 1 ACGCAGCGUGCAAACAAACGGAAGUUGGGGCGAGUGUGGAAAGCGGAAACGGAAACGAAGGAAACAUUCCCUUGUGAUAGAAAGCGGUGCAG ..(((.(((((...(.(((....))).).))...(((.(.(((.((((.((....))..(....).))))))).).)))....))).))).. ( -27.70) >consensus ACGCAGCGUGCAAUCAAACGGAAGUUAGGGCGAGCGUGGAAAGCGGAAACGGAAAC__AGAAAUCAUUCCCUUGUGAUAGAAAGCGGUGCAG ..(((.(((.(.(((((((....)))((((...((.......))(....)(....)............))))..)))).)...))).))).. (-22.10 = -21.98 + -0.13)

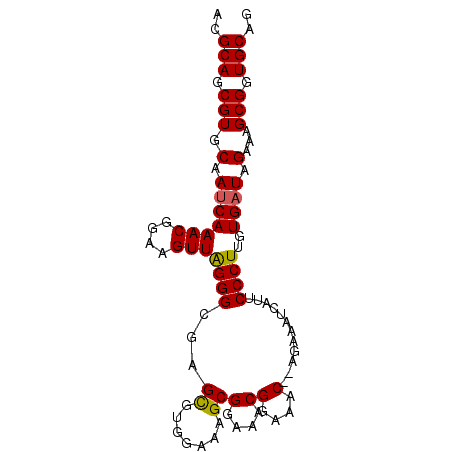
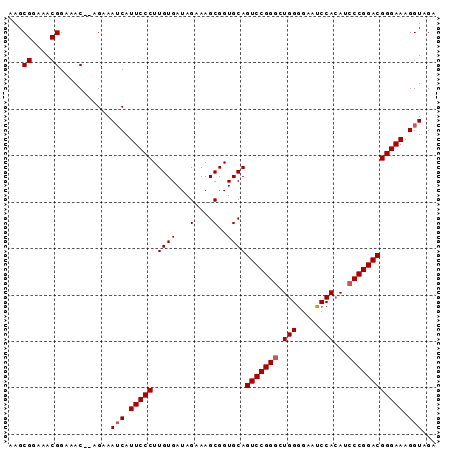
| Location | 4,345,134 – 4,345,224 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 95.05 |
| Mean single sequence MFE | -30.77 |
| Consensus MFE | -27.12 |
| Energy contribution | -27.62 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4345134 90 + 22407834 AAGCGGAAACGGAAAC--AGAAAUCAUUCCCUUGUGAUAGCAAGCGGUGCAGUCCGGGCUGGAGAAUCCACAUACCGGACGGGAAAGGUAGA ...((....)).....--....(((.(((((........(((.....))).((((((..(((.....)))....))))))))))).)))... ( -29.70) >DroSec_CAF1 13875 90 + 1 AAGCGGAAACGGAAAC--AGAAAUCAUUCCCUUGUGAUAGAAAGCGGUGCAGUCCGGGCUGGGGAAUCCACAUCCCGGACGGGAAAGGUAGA ...((....)).....--....(((.(((((((((........))))....(((((((.((((....)).)).)))))))))))).)))... ( -30.80) >DroSim_CAF1 14628 90 + 1 AAGCGGAAACGGAAAC--AGAAAUCAUUCCCUUGUGAUAGAAAGCGGUGCAGUCCGGGCUGGGGAAUCCACAUCCCGGACGGGAAAGGUAGA ...((....)).....--....(((.(((((((((........))))....(((((((.((((....)).)).)))))))))))).)))... ( -30.80) >DroYak_CAF1 14438 92 + 1 AAGCGGAAACGGAAACGAAGGAAACAUUCCCUUGUGAUAGAAAGCGGUGCAGUCCGGGCUGGGAAAUCCAGAUCCCGGACGGGAAAGGUAGA .........((....))..(....).(((((((((........))))....(((((((((((.....))))..))))))))))))....... ( -31.80) >consensus AAGCGGAAACGGAAAC__AGAAAUCAUUCCCUUGUGAUAGAAAGCGGUGCAGUCCGGGCUGGGGAAUCCACAUCCCGGACGGGAAAGGUAGA ...((....))...........(((.(((((.((((...(....)..))))(((((((.(((.....)))...)))))))))))).)))... (-27.12 = -27.62 + 0.50)

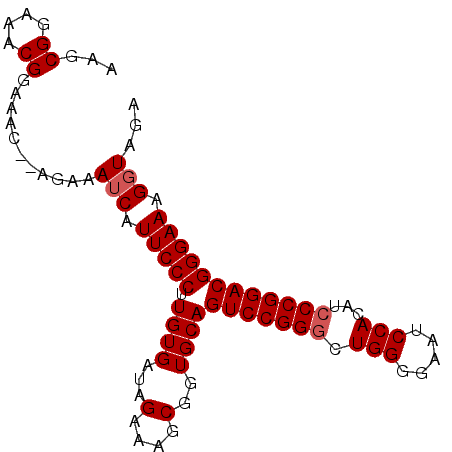
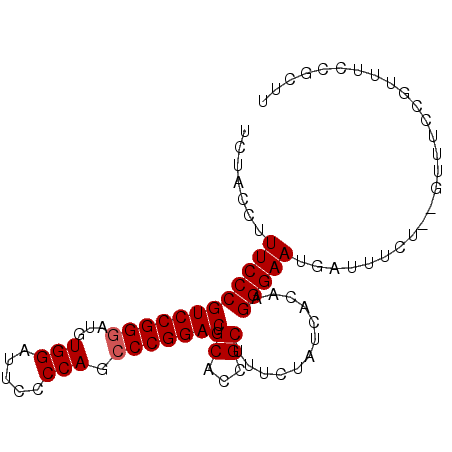
| Location | 4,345,134 – 4,345,224 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 95.05 |
| Mean single sequence MFE | -24.45 |
| Consensus MFE | -22.38 |
| Energy contribution | -22.62 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4345134 90 - 22407834 UCUACCUUUCCCGUCCGGUAUGUGGAUUCUCCAGCCCGGACUGCACCGCUUGCUAUCACAAGGGAAUGAUUUCU--GUUUCCGUUUCCGCUU .......(((((((((((....(((.....)))..)))))).(((.....)))........)))))........--................ ( -22.10) >DroSec_CAF1 13875 90 - 1 UCUACCUUUCCCGUCCGGGAUGUGGAUUCCCCAGCCCGGACUGCACCGCUUUCUAUCACAAGGGAAUGAUUUCU--GUUUCCGUUUCCGCUU ............(((((((.((.((....)))).)))))))....................(((((((......--.....))))))).... ( -24.70) >DroSim_CAF1 14628 90 - 1 UCUACCUUUCCCGUCCGGGAUGUGGAUUCCCCAGCCCGGACUGCACCGCUUUCUAUCACAAGGGAAUGAUUUCU--GUUUCCGUUUCCGCUU ............(((((((.((.((....)))).)))))))....................(((((((......--.....))))))).... ( -24.70) >DroYak_CAF1 14438 92 - 1 UCUACCUUUCCCGUCCGGGAUCUGGAUUUCCCAGCCCGGACUGCACCGCUUUCUAUCACAAGGGAAUGUUUCCUUCGUUUCCGUUUCCGCUU ............(((((((..((((.....)))))))))))....................(((((((.......))))))).......... ( -26.30) >consensus UCUACCUUUCCCGUCCGGGAUGUGGAUUCCCCAGCCCGGACUGCACCGCUUUCUAUCACAAGGGAAUGAUUUCU__GUUUCCGUUUCCGCUU .......((((((((((((...(((.....))).))))))).((...))............))))).......................... (-22.38 = -22.62 + 0.25)

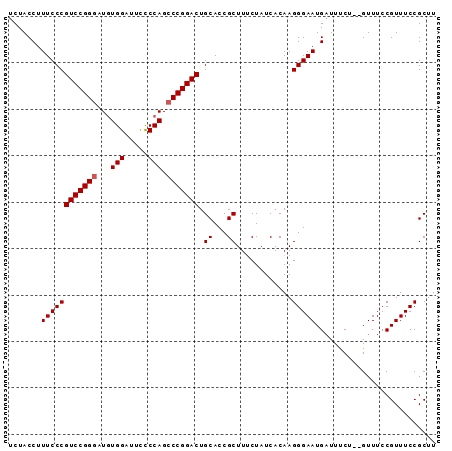
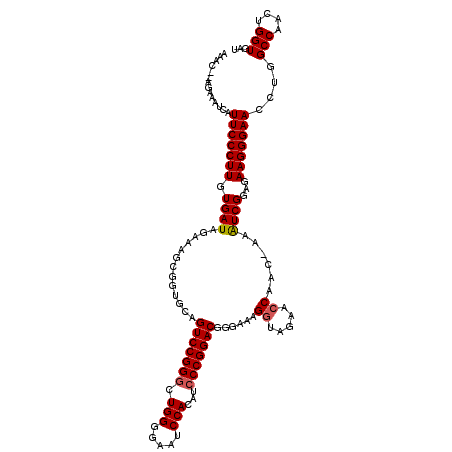
| Location | 4,345,146 – 4,345,263 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.43 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -28.52 |
| Energy contribution | -28.76 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4345146 117 + 22407834 AAAC--AGAAAUCAUUCCCUUGUGAUAGCAAGCGGUGCAGUCCGGGCUGGAGAAUCCACAUACCGGACGGGAAAGGUAGAACCAAC-AAAUCGGAGAAGGGAACCUGGCCAACUGGUGAU ...(--((..(((.(((((........(((.....))).((((((..(((.....)))....))))))))))).))).........-.....((...(((...)))..))..)))..... ( -33.20) >DroSec_CAF1 13887 118 + 1 AAAC--AGAAAUCAUUCCCUUGUGAUAGAAAGCGGUGCAGUCCGGGCUGGGGAAUCCACAUCCCGGACGGGAAAGGUAGAACCAAAAAAAUCGUAGAAGGGAACCUGGCCAACUGGUGAU ....--....(((.(((((((.((...((..........(((((((.((((....)).)).)))))))......((.....)).......)).)).)))))))....(((....)))))) ( -34.50) >DroSim_CAF1 14640 118 + 1 AAAC--AGAAAUCAUUCCCUUGUGAUAGAAAGCGGUGCAGUCCGGGCUGGGGAAUCCACAUCCCGGACGGGAAAGGUAGAACCAAAAAAAUCGGAGAAGGGAACCUGGCCAACUGGUGAU ...(--((..(((.(((((((((........))))....(((((((.((((....)).)).)))))))))))).)))...............((...(((...)))..))..)))..... ( -34.30) >DroEre_CAF1 14157 117 + 1 AAAC--AGAAAUCAUUCCCUUGUGAUAGAAAGCGGUGCAGUCCGGGCUGGGAAAUCCACAUCCCGGACAGGAAAGGUAGAACCAAC-AAAUCGGAGAAGGGAACCUGGCCAACUGGUGAU ....--....(((((......)))))......((((...(((((((.((((....)).)).))))))).((..((((....((..(-........)..))..))))..)).))))..... ( -34.30) >DroYak_CAF1 14450 119 + 1 AAACGAAGGAAACAUUCCCUUGUGAUAGAAAGCGGUGCAGUCCGGGCUGGGAAAUCCAGAUCCCGGACGGGAAAGGUAGAAGCAAC-AAGUCGGAGAAGGGAACCUGGCCAACUGGUGAU .......(....).(((((((((........))))....(((((((((((.....))))..)))))))))))).........((..-.(((.((...(((...)))..)).)))..)).. ( -35.30) >consensus AAAC__AGAAAUCAUUCCCUUGUGAUAGAAAGCGGUGCAGUCCGGGCUGGGGAAUCCACAUCCCGGACGGGAAAGGUAGAACCAAC_AAAUCGGAGAAGGGAACCUGGCCAACUGGUGAU ..............(((((((.((((.............(((((((.(((.....)))...)))))))......((.....))......))))...)))))))....(((....)))... (-28.52 = -28.76 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:48 2006