
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,342,219 – 4,342,338 |
| Length | 119 |
| Max. P | 0.999933 |

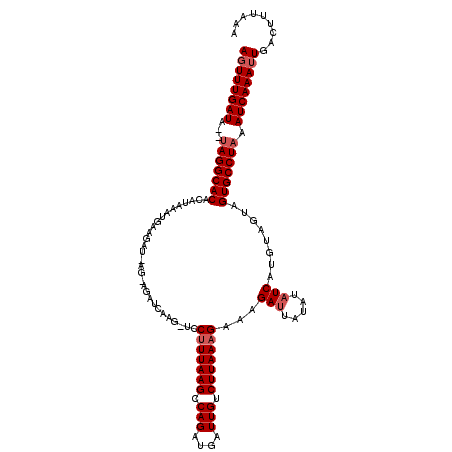
| Location | 4,342,219 – 4,342,338 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.54 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -20.37 |
| Energy contribution | -21.70 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.72 |
| SVM decision value | 4.64 |
| SVM RNA-class probability | 0.999933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
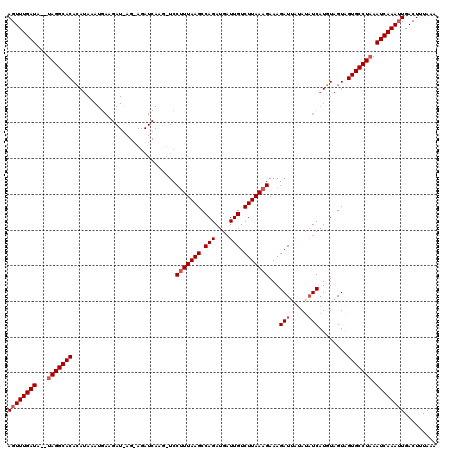
>2L_DroMel_CAF1 4342219 119 + 22407834 AGUUUGAUAUAUAGGCACAGAUAAAUGAAGAUAAGGAGAUCAAGUUCCUUUAAGCCAGAUGAUUGUCUUAAAGAAAGAUGGUAUUUCAUUUAGUGGUGCCUAAAUCAAA-UGACUUUAAG .(((((((...(((((((...(((((((((..((((((......))))))...........((..((((.....))))..)).)))))))))...))))))).))))))-)......... ( -34.10) >DroSec_CAF1 11109 94 + 1 AGUUUGAUA--UAGGCACACA------------------------UCCCUUAAGCCAGAUGCUUGUCUUAAAGAAAGAUUAUAAAUCAUGUAGUAGUGCCUCAAUCAAAUUGACUUUAAA ((((((((.--.((((((((.------------------------......((((.....))))(((((.....))))).............)).))))))..))))))))......... ( -20.90) >DroSim_CAF1 11731 118 + 1 AGUUUGAUG--UAGGCACACAUAAAUGAAGAUGAGAAGAUCAAGGUCCUUUAAGCCAGAUGAUUGUCUUAAAGAAAGAUUAAAUAUCAUGUAGUAGUGCCUAAAUCAAAUUGACUUUAAU ((((((((.--(((((((((...........(((((.(((((.(((.......)))...))))).)))))......(((.....))).....)).))))))).))))))))......... ( -30.00) >consensus AGUUUGAUA__UAGGCACACAUAAAUGAAGAU_AG_AGAUCAAG_UCCUUUAAGCCAGAUGAUUGUCUUAAAGAAAGAUUAUAUAUCAUGUAGUAGUGCCUAAAUCAAAUUGACUUUAAA ((((((((...(((((((.............................(((((((.(((....))).)))))))...(((.....)))........))))))).))))))))......... (-20.37 = -21.70 + 1.33)

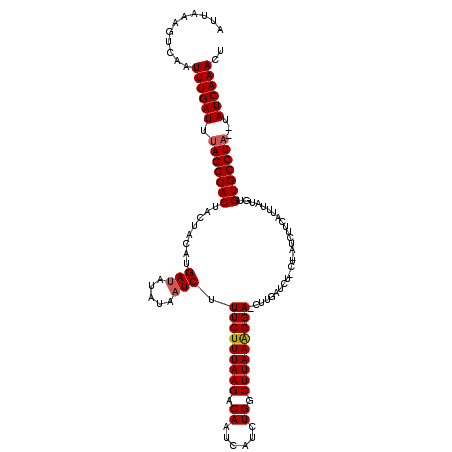
| Location | 4,342,219 – 4,342,338 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.54 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -21.19 |
| Energy contribution | -21.63 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.21 |
| Structure conservation index | 0.79 |
| SVM decision value | 4.61 |
| SVM RNA-class probability | 0.999928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
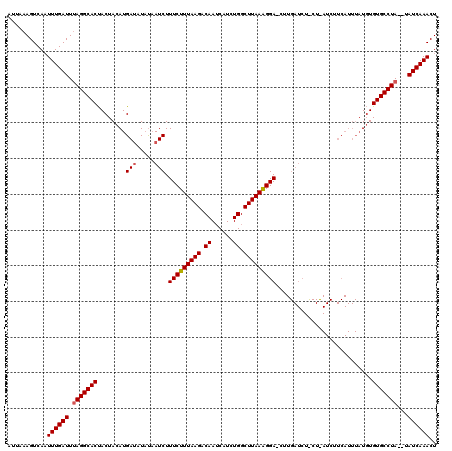
>2L_DroMel_CAF1 4342219 119 - 22407834 CUUAAAGUCA-UUUGAUUUAGGCACCACUAAAUGAAAUACCAUCUUUCUUUAAGACAAUCAUCUGGCUUAAAGGAACUUGAUCUCCUUAUCUUCAUUUAUCUGUGCCUAUAUAUCAAACU ..........-((((((.(((((((...((((((((........((((((((((.((......)).))))))))))...(((......)))))))))))...)))))))...)))))).. ( -30.20) >DroSec_CAF1 11109 94 - 1 UUUAAAGUCAAUUUGAUUGAGGCACUACUACAUGAUUUAUAAUCUUUCUUUAAGACAAGCAUCUGGCUUAAGGGA------------------------UGUGUGCCUA--UAUCAAACU ...........((((((..((((((.((.....(((.....))).(((((((((.((......)).)))))))))------------------------.)))))))).--.)))))).. ( -22.40) >DroSim_CAF1 11731 118 - 1 AUUAAAGUCAAUUUGAUUUAGGCACUACUACAUGAUAUUUAAUCUUUCUUUAAGACAAUCAUCUGGCUUAAAGGACCUUGAUCUUCUCAUCUUCAUUUAUGUGUGCCUA--CAUCAAACU ...........((((((.(((((((.((((.((((..........(((((((((.((......)).)))))))))...(((.....)))...)))).)).)))))))))--.)))))).. ( -27.60) >consensus AUUAAAGUCAAUUUGAUUUAGGCACUACUACAUGAUAUAUAAUCUUUCUUUAAGACAAUCAUCUGGCUUAAAGGA_CUUGAUCU_CU_AUCUUCAUUUAUGUGUGCCUA__UAUCAAACU ...........((((((.(((((((........(((.....))).(((((((((.((......)).)))))))))...........................)))))))...)))))).. (-21.19 = -21.63 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:43 2006