| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,335,705 – 4,335,824 |
| Length | 119 |
| Max. P | 0.549505 |

| Location | 4,335,705 – 4,335,824 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
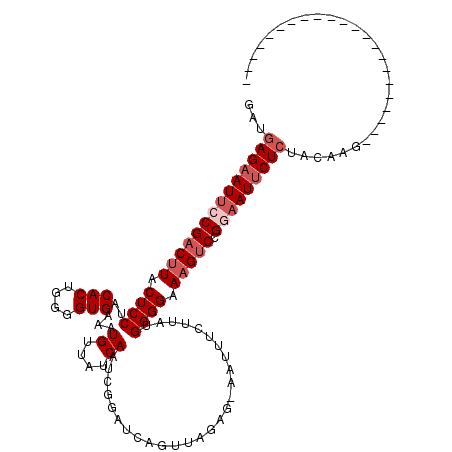
| Reading direction | forward |
| Mean pairwise identity | 81.36 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -18.08 |
| Energy contribution | -18.32 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4335705 119 + 22407834 GAGUUGUAUUGGUUCUGAGCCACUUGUAUAGAAUUCCCGCCUUUCCCCAUAAGAAAUU-CUCUAACUGAUCCGUUUGAUAACAUUUCACCCCAGUGUAGGAGUAAGUCGGAAUUCUCAUC ....(((((((((.....))))...)))))(((((((.((..((((.....(((....-.))).((((....(..(((.......)))..)))))...))))...)).)))))))..... ( -20.80) >DroSec_CAF1 4661 97 + 1 ----------------------UUUGUAGAGAAUUUCGGACUUUCCCCAUAAGAAAUU-CUCUAACUGAUCCGAUUGAUAACAUUUCACCCCAGUGUAGGAGUAAGUCGGAAUUCUCAUC ----------------------....(((((((((((((......)).....))))))-))))).....(((((((.((..(....(((....)))..)..)).)))))))......... ( -22.50) >DroSim_CAF1 5297 97 + 1 ----------------------CUUGUAGAGAAUUUCGGACUUUCCCCAUAAGAAAUU-CUCUAACUGAUCCGAUUGAUAACAUUUCACCCCAGUGUAGGAGUAAGUCGGAAUUCUCAUC ----------------------....(((((((((((((......)).....))))))-))))).....(((((((.((..(....(((....)))..)..)).)))))))......... ( -22.50) >DroEre_CAF1 4667 96 + 1 ----------------------CUUGCAGAGGAU-CCAGACUUUCCCCAUACGUUUUUUUUUCAACUGAUCCGCUUGAUAACAUUUCACCCCAGUGUAGGAGUA-GUCGGAAUUCUCAUC ----------------------......((((((-((.((((((((..(((((((........))).........(((.......))).....)))).)))).)-))))).))))))... ( -19.70) >DroYak_CAF1 4915 97 + 1 ----------------------CUUGUAGAGAAUUCCAGACUUUCCCCAUAAAGACUU-UUAUAGCUGAUUCGGCUGAUAACAUUUCACCCCAGUGUAGGAGUAAGUCGGAAUUCUCAUC ----------------------......(((((((((.(((((...........((((-(((((.(((....((.(((.......))).))))))))))))))))))))))))))))... ( -28.90) >consensus ______________________CUUGUAGAGAAUUCCAGACUUUCCCCAUAAGAAAUU_CUCUAACUGAUCCGAUUGAUAACAUUUCACCCCAGUGUAGGAGUAAGUCGGAAUUCUCAUC ............................(((((((((.(((((.(.((....................(((.....))).(((((.......))))).)).).))))))))))))))... (-18.08 = -18.32 + 0.24)

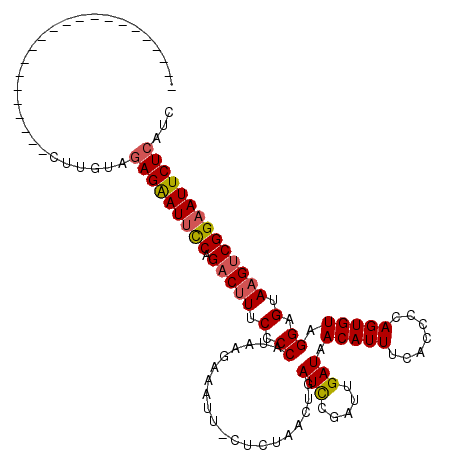
| Location | 4,335,705 – 4,335,824 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.36 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -18.66 |
| Energy contribution | -20.06 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4335705 119 - 22407834 GAUGAGAAUUCCGACUUACUCCUACACUGGGGUGAAAUGUUAUCAAACGGAUCAGUUAGAG-AAUUUCUUAUGGGGAAAGGCGGGAAUUCUAUACAAGUGGCUCAGAACCAAUACAACUC ....((((((((...(((((((......))))))).......((.(((......))).))(-..(((((.....)))))..).)))))))).......(((.......)))......... ( -25.40) >DroSec_CAF1 4661 97 - 1 GAUGAGAAUUCCGACUUACUCCUACACUGGGGUGAAAUGUUAUCAAUCGGAUCAGUUAGAG-AAUUUCUUAUGGGGAAAGUCCGAAAUUCUCUACAAA---------------------- .....((..(((((.(((((((......)))))))..((....)).))))))).((.((((-((((((....(((.....)))))))))))))))...---------------------- ( -26.40) >DroSim_CAF1 5297 97 - 1 GAUGAGAAUUCCGACUUACUCCUACACUGGGGUGAAAUGUUAUCAAUCGGAUCAGUUAGAG-AAUUUCUUAUGGGGAAAGUCCGAAAUUCUCUACAAG---------------------- .....((..(((((.(((((((......)))))))..((....)).))))))).((.((((-((((((....(((.....)))))))))))))))...---------------------- ( -26.40) >DroEre_CAF1 4667 96 - 1 GAUGAGAAUUCCGAC-UACUCCUACACUGGGGUGAAAUGUUAUCAAGCGGAUCAGUUGAAAAAAAAACGUAUGGGGAAAGUCUGG-AUCCUCUGCAAG---------------------- ...(((.((((.(((-(.(((((((.(((.(((((....)))))...)))....(((........)))))).))))..)))).))-)).)))......---------------------- ( -21.90) >DroYak_CAF1 4915 97 - 1 GAUGAGAAUUCCGACUUACUCCUACACUGGGGUGAAAUGUUAUCAGCCGAAUCAGCUAUAA-AAGUCUUUAUGGGGAAAGUCUGGAAUUCUCUACAAG---------------------- ...((((((((((((((..(((.......((.(((.......))).)).......((((((-(....))))))))))))))).)))))))))......---------------------- ( -30.00) >consensus GAUGAGAAUUCCGACUUACUCCUACACUGGGGUGAAAUGUUAUCAAUCGGAUCAGUUAGAG_AAUUUCUUAUGGGGAAAGUCCGGAAUUCUCUACAAG______________________ ...((((((((((((((.((((..(((....)))...((....))...........................)))).))))).)))))))))............................ (-18.66 = -20.06 + 1.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:39 2006