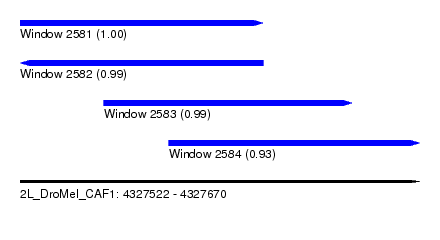
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,327,522 – 4,327,670 |
| Length | 148 |
| Max. P | 0.999883 |

| Location | 4,327,522 – 4,327,612 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 66.96 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -13.16 |
| Energy contribution | -13.60 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.34 |
| Structure conservation index | 0.53 |
| SVM decision value | 4.37 |
| SVM RNA-class probability | 0.999883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4327522 90 + 22407834 AAAUGCAACGGCAUACAUAUAUGUAGUUGUUAUAUCCCAUAUAGCA--------UCAUACUAU-CCAUA-----CUAUGUAAGCCACAUGGACGAAAGUCAAGU .........(((.((((((((((((((((((((((...))))))))--------....)))).-.))))-----.)))))).))).....(((....))).... ( -24.80) >DroSec_CAF1 81596 80 + 1 AAAGGC---------------UAUAGAUGCUAUAUCCCAUAUAGCA--------CCACAGCAU-CUAUAGAUAUCCAUGUGAGCCACAUGGACGAAAGUCAAGU .....(---------------((((((((((...............--------....)))))-)))))).....((((((...))))))(((....))).... ( -24.81) >DroSim_CAF1 85025 81 + 1 AAAGGCA--------------UAUAGAUGCUAUAUCCCAUAUAGCA--------CCAUAGUAU-CUAUAGAUAUCCAUGUGAGCCACAUGGACGAAAGUCAAGU ...(((.--------------((((((((((((.............--------..)))))))-)))))....((((((((...)))))))).....))).... ( -24.36) >DroEre_CAF1 77980 93 + 1 AAAUGCCACAGCAUACACACG----UAUGCUAUAUCGCAUAUAGCACACACGUACUAUACU--ACUGGC-----ACAUAUGAGCCACAUAGACGAAAGUCAAGU ...(((((.(((((((....)----))))))........(((((.((....)).)))))..--..))))-----)..((((.....))))(((....))).... ( -25.10) >consensus AAAGGCA______________UAUAGAUGCUAUAUCCCAUAUAGCA________CCAUACUAU_CUAUA_____CCAUGUGAGCCACAUGGACGAAAGUCAAGU ...........................((((((((...)))))))).............................((((((...))))))(((....))).... (-13.16 = -13.60 + 0.44)

| Location | 4,327,522 – 4,327,612 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 66.96 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -10.60 |
| Energy contribution | -10.16 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.60 |
| Structure conservation index | 0.38 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4327522 90 - 22407834 ACUUGACUUUCGUCCAUGUGGCUUACAUAG-----UAUGG-AUAGUAUGA--------UGCUAUAUGGGAUAUAACAACUACAUAUAUGUAUGCCGUUGCAUUU ............((((((((((...((((.-----((...-.)).)))).--------.))))))))))......((((..((((....))))..))))..... ( -21.10) >DroSec_CAF1 81596 80 - 1 ACUUGACUUUCGUCCAUGUGGCUCACAUGGAUAUCUAUAG-AUGCUGUGG--------UGCUAUAUGGGAUAUAGCAUCUAUA---------------GCCUUU ...........(((((((((...)))))))))..((((((-((((((((.--------..((.....)).)))))))))))))---------------)..... ( -30.30) >DroSim_CAF1 85025 81 - 1 ACUUGACUUUCGUCCAUGUGGCUCACAUGGAUAUCUAUAG-AUACUAUGG--------UGCUAUAUGGGAUAUAGCAUCUAUA--------------UGCCUUU ............((((((((((.(.(((((.((((....)-)))))))))--------.)))))))))).....((((....)--------------))).... ( -24.80) >DroEre_CAF1 77980 93 - 1 ACUUGACUUUCGUCUAUGUGGCUCAUAUGU-----GCCAGU--AGUAUAGUACGUGUGUGCUAUAUGCGAUAUAGCAUA----CGUGUGUAUGCUGUGGCAUUU ....(((....)))((((.....)))).((-----((((.(--((((((.((((((((((((((((...))))))))))----))))))))))))))))))).. ( -34.00) >consensus ACUUGACUUUCGUCCAUGUGGCUCACAUGG_____UAUAG_AUACUAUGG________UGCUAUAUGGGAUAUAGCAUCUAUA______________UGCAUUU ...((.((....((((((((((...(((((..............)))))..........))))))))))....))))........................... (-10.60 = -10.16 + -0.44)

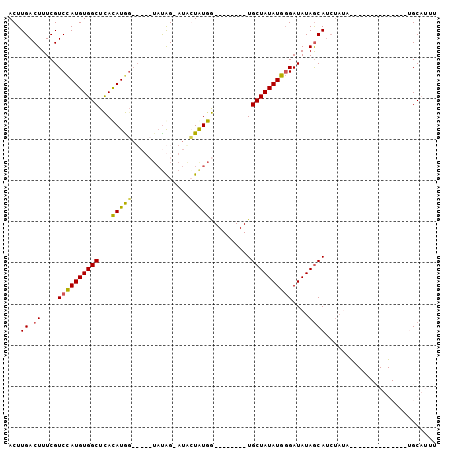
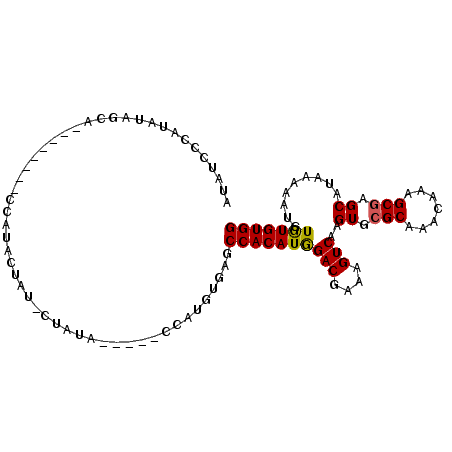
| Location | 4,327,553 – 4,327,645 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.96 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -17.25 |
| Energy contribution | -17.12 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4327553 92 + 22407834 AUAUCCCAUAUAGCA--------UCAUACUAU-CCAUA-----CUAUGUAAGCCACAUGGACGAAAGUCAAGUGCGCAAACAAAGCAAGCAUAAAAAUAUAUGUGG .....((((((((((--------(........-.....-----.(((((.....)))))(((....)))..))))((.......)).............))))))) ( -19.00) >DroSec_CAF1 81612 97 + 1 AUAUCCCAUAUAGCA--------CCACAGCAU-CUAUAGAUAUCCAUGUGAGCCACAUGGACGAAAGUCAAGUGCGCCAACAAAGCGAGCAUAAAAAUGUGUGUGG .....((((((((((--------(........-...........((((((...))))))(((....)))..))))(((........).)).........))))))) ( -24.60) >DroSim_CAF1 85042 97 + 1 AUAUCCCAUAUAGCA--------CCAUAGUAU-CUAUAGAUAUCCAUGUGAGCCACAUGGACGAAAGUCAAGUGCGCCAACAAAGCGAGCAUAAAAAUGUGUGUGG .....((((((((((--------(....((((-(....))))).((((((...))))))(((....)))..))))(((........).)).........))))))) ( -26.40) >DroEre_CAF1 78007 99 + 1 AUAUCGCAUAUAGCACACACGUACUAUACU--ACUGGC-----ACAUAUGAGCCACAUAGACGAAAGUCAAGUGCGCAAACAAGGCGAGCAUAAAAAUGUGUGUGG .............((((((((((((.....--..((((-----........))))....(((....))).)))))((.......))............))))))). ( -29.00) >consensus AUAUCCCAUAUAGCA________CCAUACUAU_CUAUA_____CCAUGUGAGCCACAUGGACGAAAGUCAAGUGCGCAAACAAAGCGAGCAUAAAAAUGUGUGUGG ....................................................((((((((((....)))..((.(((.......))).)).........))))))) (-17.25 = -17.12 + -0.12)

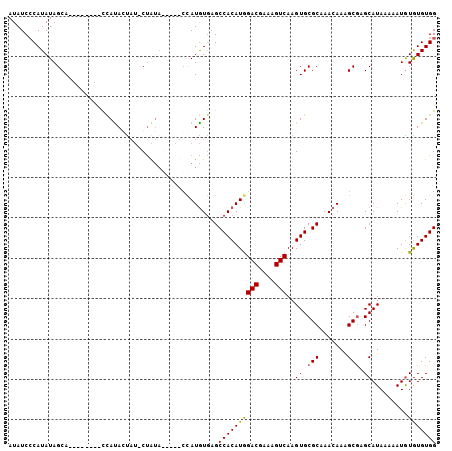
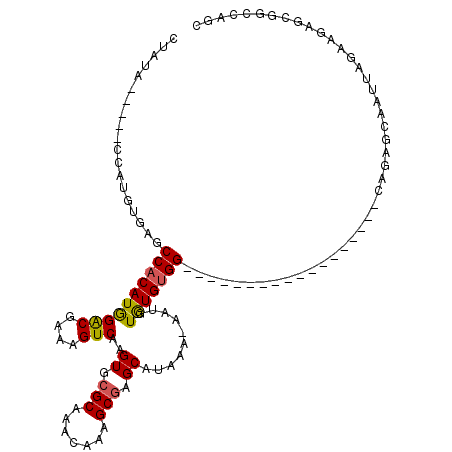
| Location | 4,327,577 – 4,327,670 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.18 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -16.85 |
| Energy contribution | -16.68 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4327577 93 + 22407834 CCAUA-----CUAUGUAAGCCACAUGGACGAAAGUCAAGUGCGCAAACAAAGCAAGCAUAAA-AAUAUAUGUGG------------------CAGAGCAAUUAGAAGAGCGGCCAGC ((...-----........(((((((((((....)))..((((((.......))..))))...-....)))))))------------------)...((..........))))..... ( -24.00) >DroSec_CAF1 81636 98 + 1 CUAUAGAUAUCCAUGUGAGCCACAUGGACGAAAGUCAAGUGCGCCAACAAAGCGAGCAUAAA-AAUGUGUGUGG------------------CAGAGCAAUUAGAAGAGCGGCCAGC .............(((..(((((...(((....)))..))).))..)))..((..(((((..-..))))).(((------------------(...((..........)).)))))) ( -24.50) >DroSim_CAF1 85066 98 + 1 CUAUAGAUAUCCAUGUGAGCCACAUGGACGAAAGUCAAGUGCGCCAACAAAGCGAGCAUAAA-AAUGUGUGUGG------------------CAGAGCAAUUAGAAGAGCAACCAGC .............(((..(((((((.(((....)))..((.(((.......))).)).....-.....))))))------------------)...))).................. ( -23.70) >DroEre_CAF1 78038 93 + 1 CUGGC-----ACAUAUGAGCCACAUAGACGAAAGUCAAGUGCGCAAACAAGGCGAGCAUAAA-AAUGUGUGUGG------------------CGGGGCAAUUAGAUGAGCGGCCAGC (((((-----........(((((((((((....)))..((.(((.......))).)).....-....)))))))------------------)...((..........)).))))). ( -32.50) >DroYak_CAF1 83920 93 + 1 CUAUA-----GCAUAUGAGCCACAUAGACGAAAGUCAAGUGCGCAAACAAAGCGAGCAUAAA-AAUGUGUGUGG------------------CAGAGCAAUUAGAAGAGCGGCCAGC .....-----((......(((((((((((....)))..((.(((.......))).)).....-....)))))))------------------)...((..........)).)).... ( -25.90) >DroAna_CAF1 56607 105 + 1 -----------CAACUGCGCC-CAUGGGCGAAAGUCAAGUGCGCCAGCACAGCGAGCAUAAAAAAUAUGUGGGGCUUGUUGUGAUGCCCAGGCCCAGCAAUUAGAGGAUCGAGCAGC -----------...(((((((-....)))....(((..((((....)))).((..(((((.....))))).(((((((..........))))))).))........)))...)))). ( -34.60) >consensus CUAUA_____CCAUGUGAGCCACAUGGACGAAAGUCAAGUGCGCAAACAAAGCGAGCAUAAA_AAUGUGUGUGG__________________CAGAGCAAUUAGAAGAGCGGCCAGC ...................((((((((((....)))..((.(((.......))).))..........)))))))........................................... (-16.85 = -16.68 + -0.17)

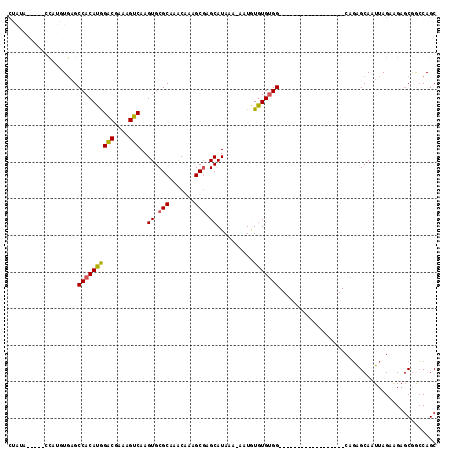
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:35 2006