
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
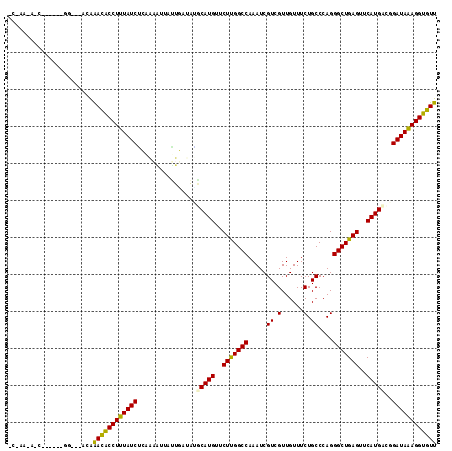
| Location | 4,316,265 – 4,316,379 |
| Length | 114 |
| Max. P | 0.980685 |

| Location | 4,316,265 – 4,316,379 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 74.39 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -28.89 |
| Energy contribution | -27.92 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
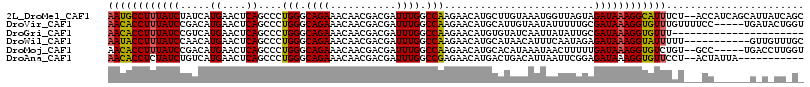
>2L_DroMel_CAF1 4316265 114 + 22407834 GCUGAUAAUGCUGAUGGU--AGAAAUGCCUUUAUCUACUAACCAUUUACAAGCAUGUUCUUGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGAUAGAUAAAGGCAUU ((.......)).......--...((((((((((((((...............((((..((..(((.....((.(......).))....)))..))..)))).)))))))))))))) ( -31.59) >DroVir_CAF1 46241 111 + 1 ACCAGUAUCA-----GGAAAACAAACACCUUUAUCGCAAAAAUAUUACAAUGCAUGUUCUUGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGUCGGAUAAAGGUGUU .((.......-----))......(((((((((((((((............)))(((..((..(((.....((.(......).))....)))..))..)))....)))))))))))) ( -30.10) >DroGri_CAF1 65327 94 + 1 ----------------------AAACACCUUUAUCGCAAUAUAAUUGAUACACAUGUUCUUGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGACGGAUAAAGGUGUU ----------------------.((((((((((((.((((...)))).....((((..((..(((.....((.(......).))....)))..))..))))...)))))))))))) ( -30.90) >DroWil_CAF1 81454 105 + 1 GCAAACAAC-----------AAAAAUACCUUUAUCUCUAUUGAAAUGUUAUGCAUGUUCUUGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGUUGGAUAAAGGUAUU .........-----------...((((((((((((((....))........(((((..((..(((.....((.(......).))....)))..))..)))))..)))))))))))) ( -29.20) >DroMoj_CAF1 51172 109 + 1 ACCAAGGUCA-----GGC--ACAGACACCUUUAUCAAAAAGUUAUUUAUGUGCAUGUUCUUGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGUCGGAUAAAGGUGUU ..........-----...--...((((((((((((..(((....)))....(((((..((..(((.....((.(......).))....)))..))..)))))..)))))))))))) ( -30.60) >DroAna_CAF1 47469 103 + 1 -----------UAAUAGU--AGGAACACCUUUAUCUCCGAAUUAAUGUCAGUCAUGUUCUCGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGACAGAUAGAGGUGUU -----------.......--...(((((((((((((..((.......)).((((((..(((((((.....((.(......).))....)))))))..))))))))))))))))))) ( -38.30) >consensus _C_AA_A_C______GG___ACAAACACCUUUAUCUCAAAAUUAUUGAUAUGCAUGUUCUUGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGACGGAUAAAGGUGUU .......................((((((((((((................(((((..(((((((.....((.(......).))....)))))))..)))))..)))))))))))) (-28.89 = -27.92 + -0.97)


| Location | 4,316,265 – 4,316,379 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 74.39 |
| Mean single sequence MFE | -24.22 |
| Consensus MFE | -19.98 |
| Energy contribution | -19.45 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4316265 114 - 22407834 AAUGCCUUUAUCUAUCAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCAAGAACAUGCUUGUAAAUGGUUAGUAGAUAAAGGCAUUUCU--ACCAUCAGCAUUAUCAGC (((((((((((((((..........(((...))).........(((.(((((..((((......))))))))).))).)))))))))))))))...--.................. ( -28.70) >DroVir_CAF1 46241 111 - 1 AACACCUUUAUCCGACAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCAAGAACAUGCAUUGUAAUAUUUUUGCGAUAAAGGUGUUUGUUUUCC-----UGAUACUGGU ((((((((((((.(.((((....((....))(((.(((.(......).))).))).....)))))...((((.....))))))))))))))))......((-----.......)). ( -24.70) >DroGri_CAF1 65327 94 - 1 AACACCUUUAUCCGUCAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCAAGAACAUGUGUAUCAAUUAUAUUGCGAUAAAGGUGUUU---------------------- ((((((((((((((((.((....))(((...))).........))))................(((((.....)))))...)))))))))))).---------------------- ( -23.80) >DroWil_CAF1 81454 105 - 1 AAUACCUUUAUCCAACAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCAAGAACAUGCAUAACAUUUCAAUAGAGAUAAAGGUAUUUUU-----------GUUGUUUGC (((((((((((((................(((.((((...........)))).))).(((.(((.....))))))....).))))))))))))...-----------......... ( -20.50) >DroMoj_CAF1 51172 109 - 1 AACACCUUUAUCCGACAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCAAGAACAUGCACAUAAAUAACUUUUUGAUAAAGGUGUCUGU--GCC-----UGACCUUGGU .(((((((((((.(.((((....((....))(((.(((.(......).))).))).....)))))....(((......))))))))))))))....--(((-----.......))) ( -21.60) >DroAna_CAF1 47469 103 - 1 AACACCUCUAUCUGUCAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCGAGAACAUGACUGACAUUAAUUCGGAGAUAAAGGUGUUCCU--ACUAUUA----------- (((((((.(((((.(((((..(((.(((.......................))).)))..)))))((((.......))))))))).)))))))...--.......----------- ( -26.00) >consensus AACACCUUUAUCCGACAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCAAGAACAUGCAUAUCAAUAAUUUUGAGAUAAAGGUGUUUCU___CC______G_U_UU_G_ ((((((((((((.....((....))....(((.((((...........)))).))).........................))))))))))))....................... (-19.98 = -19.45 + -0.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:29 2006