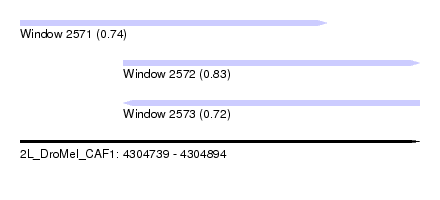
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,304,739 – 4,304,894 |
| Length | 155 |
| Max. P | 0.834905 |

| Location | 4,304,739 – 4,304,858 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.11 |
| Mean single sequence MFE | -36.68 |
| Consensus MFE | -34.00 |
| Energy contribution | -33.42 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
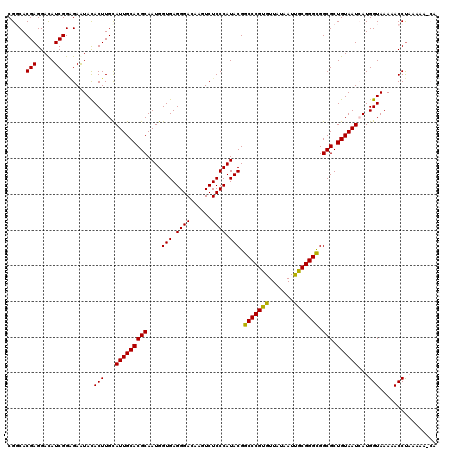
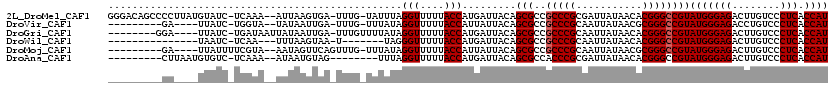
>2L_DroMel_CAF1 4304739 119 + 22407834 CGGCACGAGGACAUCGGAGAGUACACUUGCAUUGCACGCAAUGGUGAGGGACAAGUCUCCCAUACGGCCCGUGUUAUAAUCGCGGGCGGCGCUGUAAUCAUGGUAAAAACCUAAAUA-CA .((..(((.....))).....(((...((.(((((((((....(((.((((......)))).))).(((((((.......))))))).))).))))))))..)))....))......-.. ( -36.80) >DroVir_CAF1 36344 119 + 1 CGGCACGAGGAUAUCGGAGAAUACACUUGCAUUGCACGCAAUGGUGAGGGACAGGUCUCCCAUACGGCCCGCGUUAUAAUUGCGGGCGGCGCUGUAAUAAUGGUAAAAACCUAUAAA-CA ..((.(((.....)))(((......)))))(((((((((....(((.((((......)))).))).(((((((.......))))))).))).))))))...(((....)))......-.. ( -36.70) >DroGri_CAF1 50529 120 + 1 CGGCACGAGGACAUCGGCGAAUACACUUGUAUUGCACGCAAUGGUGAGGGACAAGUCUCCCAUACGGCCCGUGUUAUAAUUGCGGGCGGCGCUGUAAUCAUGGUAAAAACCUAUAAAACA ..((.(((.....)))))...(((...((.(((((((((....(((.((((......)))).))).(((((((.......))))))).))).))))))))..)))............... ( -36.50) >DroWil_CAF1 69195 114 + 1 CGCCACGAGGAUAUCGGGGAAUAUACUUGCAUUGCACGCAAUGGUGAGGGACAAGUCUCCCAUACGGCCCGUGUUAUAAUUGCGGGCGGCGCUGUAAUCAUGGUAAAAACCCUA------ .....(((.....)))(((....((((((.(((((((((....(((.((((......)))).))).(((((((.......))))))).))).)))))))).))))....)))..------ ( -38.00) >DroMoj_CAF1 42815 119 + 1 CGCCACGAGGACAUCGGAGAAUACACUUGCAUUGCACGCAAUGGUGAGGGACAAGUCUCCCAUACGGCCCGCGUUAUAAUUGCGGGCGGCGCUGUAAUAAUGGUAAAAACCUAUAAA-CA .(((((((.....)))..............(((((((((....(((.((((......)))).))).(((((((.......))))))).))).))))))..)))).............-.. ( -37.30) >DroAna_CAF1 39217 115 + 1 CGGCACGAGGACAUCGGAGAGUACACUUGCAUUGCACGCAAUGGUGAGGGACAAGUCUCCCAUACGGCCCGUGUUAUAAUCGCGGGUGGCGCUGUAAUCAUGGUAAAAACCUAAA----- .((..(((.....))).....(((...((.(((((((((....(((.((((......)))).))).(((((((.......))))))).))).))))))))..)))....))....----- ( -34.80) >consensus CGGCACGAGGACAUCGGAGAAUACACUUGCAUUGCACGCAAUGGUGAGGGACAAGUCUCCCAUACGGCCCGUGUUAUAAUUGCGGGCGGCGCUGUAAUCAUGGUAAAAACCUAAAAA_CA .....(((.....)))........(((...(((((((((....(((.((((......)))).))).(((((((.......))))))).))).))))))...)))................ (-34.00 = -33.42 + -0.58)

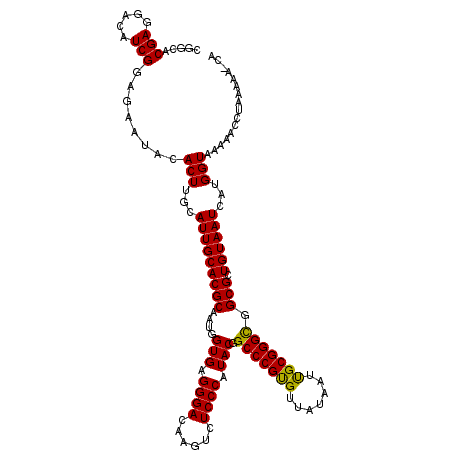
| Location | 4,304,779 – 4,304,894 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.84 |
| Mean single sequence MFE | -27.05 |
| Consensus MFE | -25.26 |
| Energy contribution | -24.68 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4304779 115 + 22407834 AUGGUGAGGGACAAGUCUCCCAUACGGCCCGUGUUAUAAUCGCGGGCGGCGCUGUAAUCAUGGUAAAAACCUAAAUA-CAAA-UCACUUAAU--UUUGA-GAUACAUAAGGGGCUGUCCC .......((((((.(((((.....(.(((((((.......))))))).)...((((.((..(((....)))......-((((-.........--)))).-))))))...))))))))))) ( -34.60) >DroVir_CAF1 36384 102 + 1 AUGGUGAGGGACAGGUCUCCCAUACGGCCCGCGUUAUAAUUGCGGGCGGCGCUGUAAUAAUGGUAAAAACCUAUAAA-CAAA-UCAAUUAUA--UACCA-GAUAA----UC--------- .(((((.((((......))))...(.(((((((.......))))))).)...(((......(((....))).....)-))..-.........--)))))-.....----..--------- ( -27.20) >DroGri_CAF1 50569 106 + 1 AUGGUGAGGGACAAGUCUCCCAUACGGCCCGUGUUAUAAUUGCGGGCGGCGCUGUAAUCAUGGUAAAAACCUAUAAAACAAA-UCAAUUAUAAUUAUCA-GAUAA----UCC-------- ((((((.((((......))))...(.(((((((.......))))))).))))))).(((.((((((.....(((((......-....))))).))))))-)))..----...-------- ( -24.30) >DroWil_CAF1 69235 93 + 1 AUGGUGAGGGACAAGUCUCCCAUACGGCCCGUGUUAUAAUUGCGGGCGGCGCUGUAAUCAUGGUAAAAACCCUA-------A-UUACUUAAA---UUGA-GAUUA--------------- ..((((.((((......))))...(.(((((((.......))))))).)))))(((((...(((....)))...-------)-)))).....---....-.....--------------- ( -25.30) >DroMoj_CAF1 42855 104 + 1 AUGGUGAGGGACAAGUCUCCCAUACGGCCCGCGUUAUAAUUGCGGGCGGCGCUGUAAUAAUGGUAAAAACCUAUAAA-CAAACUGAACUAUU--UACGAAAAUAA----UC--------- ((((((.((((......))))...(.(((((((.......))))))).)))))))......(((....)))......-..............--...........----..--------- ( -26.00) >DroAna_CAF1 39257 100 + 1 AUGGUGAGGGACAAGUCUCCCAUACGGCCCGUGUUAUAAUCGCGGGUGGCGCUGUAAUCAUGGUAAAAACCUAAA--------CUACAUUAU--UUUGA-GACACAUUAAG--------- ...(((.((((......))))...(.(((((((.......))))))).)...((((.....(((....)))....--------.))))....--.....-..)))......--------- ( -24.90) >consensus AUGGUGAGGGACAAGUCUCCCAUACGGCCCGUGUUAUAAUUGCGGGCGGCGCUGUAAUCAUGGUAAAAACCUAAAAA_CAAA_UCAAUUAAA__UAUGA_GAUAA____UC_________ ((((((.((((......))))...(.(((((((.......))))))).)))))))......(((....)))................................................. (-25.26 = -24.68 + -0.58)

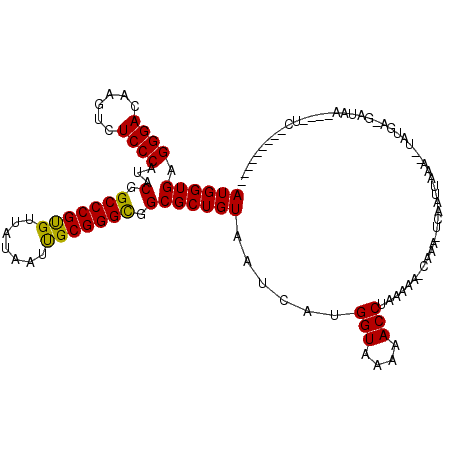
| Location | 4,304,779 – 4,304,894 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.84 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -17.78 |
| Energy contribution | -17.95 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
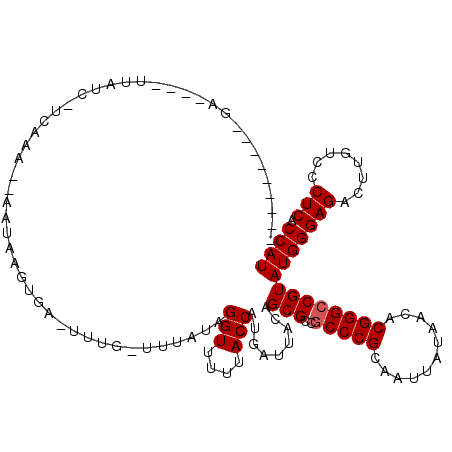
>2L_DroMel_CAF1 4304779 115 - 22407834 GGGACAGCCCCUUAUGUAUC-UCAAA--AUUAAGUGA-UUUG-UAUUUAGGUUUUUACCAUGAUUACAGCGCCGCCCGCGAUUAUAACACGGGCCGUAUGGGAGACUUGUCCCUCACCAU (((((((((((.((((....-.....--.....(((.-..((-((.((((((....))).))).)))).))).(((((.(.......).))))))))).))).).).))))))....... ( -35.20) >DroVir_CAF1 36384 102 - 1 ---------GA----UUAUC-UGGUA--UAUAAUUGA-UUUG-UUUAUAGGUUUUUACCAUUAUUACAGCGCCGCCCGCAAUUAUAACGCGGGCCGUAUGGGAGACCUGUCCCUCACCAU ---------..----.....-((((.--.((((....-.)))-)..(((((((((..((((.......(((..((((((.........)))))))))))))))))))))).....)))). ( -25.91) >DroGri_CAF1 50569 106 - 1 --------GGA----UUAUC-UGAUAAUUAUAAUUGA-UUUGUUUUAUAGGUUUUUACCAUGAUUACAGCGCCGCCCGCAAUUAUAACACGGGCCGUAUGGGAGACUUGUCCCUCACCAU --------((.----....(-((.(((((((...(((-......)))..(((....))))))))))))).((.(((((...........))))).))..((((......))))...)).. ( -22.70) >DroWil_CAF1 69235 93 - 1 ---------------UAAUC-UCAA---UUUAAGUAA-U-------UAGGGUUUUUACCAUGAUUACAGCGCCGCCCGCAAUUAUAACACGGGCCGUAUGGGAGACUUGUCCCUCACCAU ---------------.....-....---.....((((-(-------((.(((....))).)))))))...((.(((((...........))))).))(((((((........))).)))) ( -24.90) >DroMoj_CAF1 42855 104 - 1 ---------GA----UUAUUUUCGUA--AAUAGUUCAGUUUG-UUUAUAGGUUUUUACCAUUAUUACAGCGCCGCCCGCAAUUAUAACGCGGGCCGUAUGGGAGACUUGUCCCUCACCAU ---------..----........(((--(((((......)))-))))).(((....)))...........((.((((((.........)))))).))(((((((........))).)))) ( -25.90) >DroAna_CAF1 39257 100 - 1 ---------CUUAAUGUGUC-UCAAA--AUAAUGUAG--------UUUAGGUUUUUACCAUGAUUACAGCGCCACCCGCGAUUAUAACACGGGCCGUAUGGGAGACUUGUCCCUCACCAU ---------......((.((-(((..--....(((((--------((..(((....)))..)))))))(((...((((.(.......).)))).))).))))).)).............. ( -23.20) >consensus _________GA____UUAUC_UCAAA__AAUAAGUGA_UUUG_UUUAUAGGUUUUUACCAUGAUUACAGCGCCGCCCGCAAUUAUAACACGGGCCGUAUGGGAGACUUGUCCCUCACCAU .................................................(((....))).........(((..(((((...........))))))))(((((((........))).)))) (-17.78 = -17.95 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:25 2006