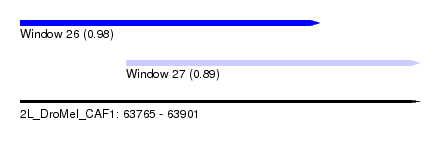
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 63,765 – 63,901 |
| Length | 136 |
| Max. P | 0.979805 |

| Location | 63,765 – 63,867 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.82 |
| Mean single sequence MFE | -27.86 |
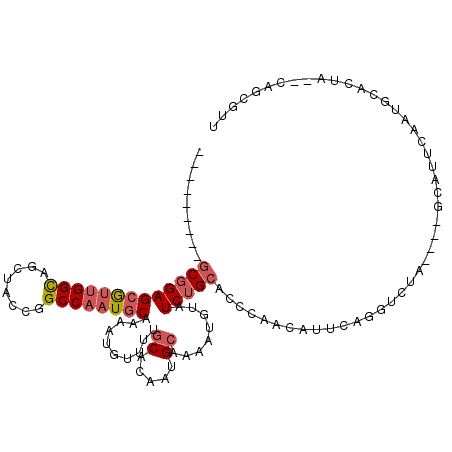
| Consensus MFE | -16.76 |
| Energy contribution | -17.48 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
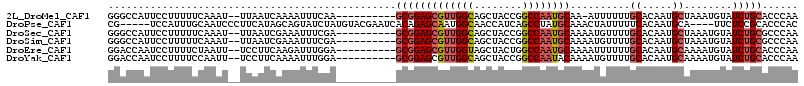
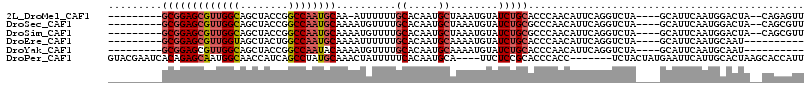
>2L_DroMel_CAF1 63765 102 + 22407834 GGGCCAUUCCUUUUUCAAAU--UUAAUCAAAAUUUCAA----------GCGGAGCGUUGGCAGCUACCGGCCAAUGCAA-AUUUUUUGCACAAUGCUAAAUGUAUCUGCACCCAA (((.............((((--((.....))))))...----------(((((((((((((........))))))))..-((((...((.....)).))))...))))).))).. ( -25.90) >DroPse_CAF1 49388 106 + 1 CG-----UCCAUUUGCAAUCCCUUCAUAGCAGUAUCUAUGUACGAAUCACAGAGCAAUGGCAACCAUCAGCCUAUGCAAACUAUUUUUCACAAUGCA----UUCUCCGCACCCAC .(-----(.....((((.((....(((((......)))))...))........(((..(((........)))..)))................))))----......))...... ( -14.80) >DroSec_CAF1 36470 103 + 1 GGGCCAUUCCUUUUUCAAAU--UUAAUCGAAAUUUCGA----------GCGGAGCGUUGGCAGCUACCGGCCAAUGCAAAAUGUUUUGCACAAUGCUAAAUGUAUCUGCGCCCAA ((((................--....((((....))))----------(((((((((((((........))))))))...((((((.((.....)).)))))).))))))))).. ( -34.80) >DroSim_CAF1 38771 103 + 1 GGGCCAUUCCUUUUUCAAAU--UUAAUCGAAAUUUCGA----------GCGGAGCGUUGGCAGCUACCGGCCAAUGCAAAAUGUUUUGCACAAUGCUAAAUGUAUCUGCGCCCAA ((((................--....((((....))))----------(((((((((((((........))))))))...((((((.((.....)).)))))).))))))))).. ( -34.80) >DroEre_CAF1 42885 103 + 1 GGACCAAUCCUUUUCUAAUU--UCCUUCAAGAUUUGGA----------GCGGAGCGUUGGUAGCUACUGGCCAAUGCAAAAUUUUUUGCACAAUGCAAAAUGUAUCUGCACCCAA ((.((((..(((........--......)))..)))).----------(((((((((((((........))))))))......((((((.....))))))....)))))..)).. ( -29.04) >DroYak_CAF1 36339 103 + 1 GGACCAAUCCUUUUCCAAUU--UCCUUCAAAAUUUGGA----------GCGGAGCGUUGGCAGCUACCGGCCAAUACAAAAUGUUUUGCACAAUGCAAAAUGUAUCUGCACCCAA ((...........(((((..--...........)))))----------((((((..(((((........)))))..)...(((((((((.....))))))))).)))))..)).. ( -27.82) >consensus GGGCCAUUCCUUUUUCAAAU__UUAAUCAAAAUUUCGA__________GCGGAGCGUUGGCAGCUACCGGCCAAUGCAAAAUGUUUUGCACAAUGCAAAAUGUAUCUGCACCCAA ................................................(((((((((((((........))))))))..........((.....))........)))))...... (-16.76 = -17.48 + 0.72)
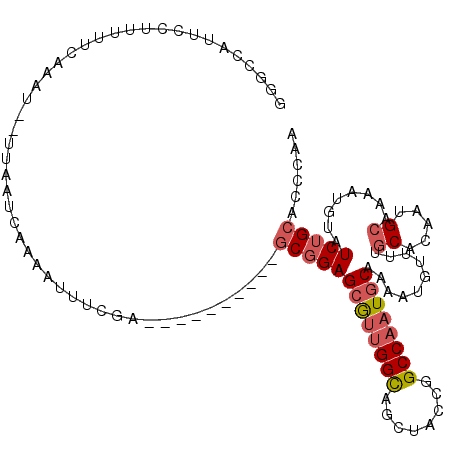
| Location | 63,801 – 63,901 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.69 |
| Mean single sequence MFE | -25.77 |
| Consensus MFE | -16.76 |
| Energy contribution | -17.48 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 63801 100 + 22407834 ---------GCGGAGCGUUGGCAGCUACCGGCCAAUGCAA-AUUUUUUGCACAAUGCUAAAUGUAUCUGCACCCAACAUUCAGGUCUA----GCAUUCAAUGGACUA--CAGAGUU ---------(((((((((((((........))))))))..-((((...((.....)).))))...))))).......((((.((((((----........)))))).--..)))). ( -27.80) >DroSec_CAF1 36506 101 + 1 ---------GCGGAGCGUUGGCAGCUACCGGCCAAUGCAAAAUGUUUUGCACAAUGCUAAAUGUAUCUGCGCCCAACAUUCAGGUCUA----GCAUUCAAUGGACUA--CAGCGUU ---------((...((((((((........))))))))..((((((..((.((((((.....)))).)).))..))))))..((((((----........)))))).--..))... ( -30.40) >DroSim_CAF1 38807 101 + 1 ---------GCGGAGCGUUGGCAGCUACCGGCCAAUGCAAAAUGUUUUGCACAAUGCUAAAUGUAUCUGCGCCCAACAUUCAGGUCUA----GCAUUCAAUGGACUA--CAGCGUU ---------((...((((((((........))))))))..((((((..((.((((((.....)))).)).))..))))))..((((((----........)))))).--..))... ( -30.40) >DroEre_CAF1 42921 93 + 1 ---------GCGGAGCGUUGGUAGCUACUGGCCAAUGCAAAAUUUUUUGCACAAUGCAAAAUGUAUCUGCACCCAACAUUCAGGUCUA----GCAUUCAAUGCAAU---------- ---------(((((((((((((........))))))))......((((((.....))))))....)))))..((........))....----((((...))))...---------- ( -26.00) >DroYak_CAF1 36375 93 + 1 ---------GCGGAGCGUUGGCAGCUACCGGCCAAUACAAAAUGUUUUGCACAAUGCAAAAUGUAUCUGCACCCAACAUUCAGGUCUA----GCAUUCAAUGCAAU---------- ---------((((((..(((((........)))))..)...(((((((((.....))))))))).)))))..((........))....----((((...))))...---------- ( -24.70) >DroPer_CAF1 49280 105 + 1 GUACGAAUCACAGAGCAAUGGCAACCAUCAGCCUAUGCAAACUAUUUUUCACAAUGCA----UUCUCCGCACCCACC-------UCUACUAUGAAUUCAUUGCACUAAGCACCAUU ....((((((.((.(((..(((........)))..)))................(((.----......)))......-------....)).)).))))..(((.....)))..... ( -15.30) >consensus _________GCGGAGCGUUGGCAGCUACCGGCCAAUGCAAAAUGUUUUGCACAAUGCAAAAUGUAUCUGCACCCAACAUUCAGGUCUA____GCAUUCAAUGCACUA__CAGCGUU .........(((((((((((((........))))))))..........((.....))........))))).............................................. (-16.76 = -17.48 + 0.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:23:20 2006