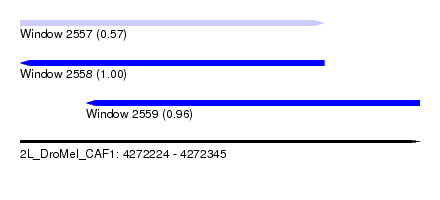
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,272,224 – 4,272,345 |
| Length | 121 |
| Max. P | 0.998815 |

| Location | 4,272,224 – 4,272,316 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 96.48 |
| Mean single sequence MFE | -15.54 |
| Consensus MFE | -13.27 |
| Energy contribution | -13.91 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
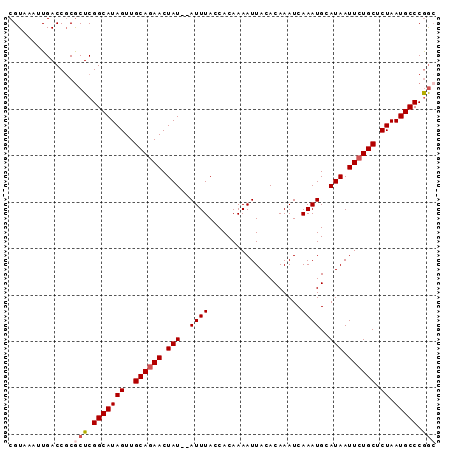
>2L_DroMel_CAF1 4272224 92 + 22407834 CGUAAAUUGACCGCGCUCGGCAUAGUUGCAGAACUAUAUAUUUACCACAAAAUUACACAAAUCAAAUGCAUAAUUCUGCUCUAAUGCCCGGC ..............(((.(((((((..((((((.(((.(((((....................))))).))).)))))).)).))))).))) ( -19.85) >DroSec_CAF1 26838 90 + 1 CGUAAAUUGACCGCGCUCGGCAUAGUUGCAGAACUAU--AUUUACCACAAAAUUACACAAAUCAAAUGCAUAAUUCUGCUCUAAUGCCCGGG ..........(((.....(((((((..((((((.(((--..............................))).)))))).)).)))))))). ( -16.01) >DroSim_CAF1 28066 90 + 1 CGUAAAUUGACCGCGCUCGGCAUAGUUGCAGAACUAU--AUUUACCACAAAAUUACACAAAUCAAAUGCAUAAUUCUGCUCUAAUGCCCGGC ..............(((.(((((((..((((((.(((--..............................))).)))))).)).))))).))) ( -17.21) >DroEre_CAF1 22378 90 + 1 CGUAAAUUGACCUCACUCGGCAUAGUUGCACAACUAU--AUUUACCACAAAAUUACACAAAUCAAAUGCAUAAUUCUGCUCUAAUGCCCAGC ...............((.(((((((((....))))))--............................(((......)))......))).)). ( -11.00) >DroYak_CAF1 26985 90 + 1 CGUAAAUUGACCGCGCUCGGCAUAGUUGCAGAACUAU--AUUUACCACAAAAUUACACAAAUCAAAUGCAUAAUUCUGCUCUAAUGCCCGAC ..................(((((((..((((((.(((--..............................))).)))))).)).))))).... ( -13.61) >consensus CGUAAAUUGACCGCGCUCGGCAUAGUUGCAGAACUAU__AUUUACCACAAAAUUACACAAAUCAAAUGCAUAAUUCUGCUCUAAUGCCCGGC ..............(((.(((((((..((((((.(((..((((....................))))..))).)))))).)).))))).))) (-13.27 = -13.91 + 0.64)


| Location | 4,272,224 – 4,272,316 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 96.48 |
| Mean single sequence MFE | -26.16 |
| Consensus MFE | -23.68 |
| Energy contribution | -23.40 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
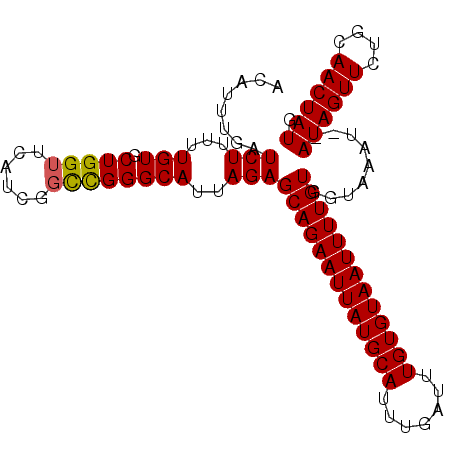
>2L_DroMel_CAF1 4272224 92 - 22407834 GCCGGGCAUUAGAGCAGAAUUAUGCAUUUGAUUUGUGUAAUUUUGUGGUAAAUAUAUAGUUCUGCAACUAUGCCGAGCGCGGUCAAUUUACG ((((.((.(((..(((((((((((((.......)))))))))))))..)))...(((((((....)))))))....)).))))......... ( -27.00) >DroSec_CAF1 26838 90 - 1 CCCGGGCAUUAGAGCAGAAUUAUGCAUUUGAUUUGUGUAAUUUUGUGGUAAAU--AUAGUUCUGCAACUAUGCCGAGCGCGGUCAAUUUACG .(((.((......(((((((((((((.......)))))))))))))(((....--((((((....)))))))))..)).))).......... ( -26.10) >DroSim_CAF1 28066 90 - 1 GCCGGGCAUUAGAGCAGAAUUAUGCAUUUGAUUUGUGUAAUUUUGUGGUAAAU--AUAGUUCUGCAACUAUGCCGAGCGCGGUCAAUUUACG ((((.((......(((((((((((((.......)))))))))))))(((....--((((((....)))))))))..)).))))......... ( -27.30) >DroEre_CAF1 22378 90 - 1 GCUGGGCAUUAGAGCAGAAUUAUGCAUUUGAUUUGUGUAAUUUUGUGGUAAAU--AUAGUUGUGCAACUAUGCCGAGUGAGGUCAAUUUACG (((.(((.(((..(((((((((((((.......)))))))))))))..)))..--((((((....))))))))).))).............. ( -26.60) >DroYak_CAF1 26985 90 - 1 GUCGGGCAUUAGAGCAGAAUUAUGCAUUUGAUUUGUGUAAUUUUGUGGUAAAU--AUAGUUCUGCAACUAUGCCGAGCGCGGUCAAUUUACG ((((.((......(((((((((((((.......)))))))))))))(((....--((((((....)))))))))..)).))).)........ ( -23.80) >consensus GCCGGGCAUUAGAGCAGAAUUAUGCAUUUGAUUUGUGUAAUUUUGUGGUAAAU__AUAGUUCUGCAACUAUGCCGAGCGCGGUCAAUUUACG .(((.((......(((((((((((((.......)))))))))))))(((......((((((....)))))))))..)).))).......... (-23.68 = -23.40 + -0.28)



| Location | 4,272,244 – 4,272,345 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 96.79 |
| Mean single sequence MFE | -23.68 |
| Consensus MFE | -22.06 |
| Energy contribution | -21.94 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4272244 101 - 22407834 ACAUUUGAUCUUUUUGUGCUGGUUCAUCGGCCGGGCAUUAGAGCAGAAUUAUGCAUUUGAUUUGUGUAAUUUUGUGGUAAAUAUAUAGUUCUGCAACUAUG ..(((((.(((...(((.(((((......))))))))..)))(((((((((((((.......)))))))))))))..))))).(((((((....))))))) ( -24.90) >DroSec_CAF1 26858 99 - 1 ACAUUUGAUCUUUUUGUGCUGGUUCAUCGCCCGGGCAUUAGAGCAGAAUUAUGCAUUUGAUUUGUGUAAUUUUGUGGUAAAU--AUAGUUCUGCAACUAUG ........(((...(((.(.(((.....))).).)))..)))(((((((((((((.......)))))))))))))......(--((((((....))))))) ( -23.30) >DroSim_CAF1 28086 99 - 1 ACAUUUGAUCUUUUUGUGCUGGUUCAUCGGCCGGGCAUUAGAGCAGAAUUAUGCAUUUGAUUUGUGUAAUUUUGUGGUAAAU--AUAGUUCUGCAACUAUG ........(((...(((.(((((......))))))))..)))(((((((((((((.......)))))))))))))......(--((((((....))))))) ( -24.60) >DroEre_CAF1 22398 99 - 1 ACAUUUGAUCUUUUUGUGCUGGUUCAUCAGCUGGGCAUUAGAGCAGAAUUAUGCAUUUGAUUUGUGUAAUUUUGUGGUAAAU--AUAGUUGUGCAACUAUG ...(((((((((.....(((((....))))).))).))))))(((((((((((((.......)))))))))))))......(--((((((....))))))) ( -23.70) >DroYak_CAF1 27005 99 - 1 ACAUUUGAUCUUUUUGUGCUGGUUCAUUGGUCGGGCAUUAGAGCAGAAUUAUGCAUUUGAUUUGUGUAAUUUUGUGGUAAAU--AUAGUUCUGCAACUAUG ........(((....(((((.(.........).))))).)))(((((((((((((.......)))))))))))))......(--((((((....))))))) ( -21.90) >consensus ACAUUUGAUCUUUUUGUGCUGGUUCAUCGGCCGGGCAUUAGAGCAGAAUUAUGCAUUUGAUUUGUGUAAUUUUGUGGUAAAU__AUAGUUCUGCAACUAUG ........(((...(((.(((((......))))))))..)))(((((((((((((.......))))))))))))).........((((((....)))))). (-22.06 = -21.94 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:11 2006