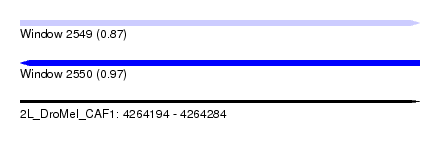
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,264,194 – 4,264,284 |
| Length | 90 |
| Max. P | 0.973333 |

| Location | 4,264,194 – 4,264,284 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -19.32 |
| Energy contribution | -19.56 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.874206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4264194 90 + 22407834 AAACCCGCACAAAGCCGGAAAACGUGGCAAUUGUGUGCUGGCCCAGCAUCUCAAACCCAUGUCCUCCUUCUACAUGGCACCUGGAGAAAA ......((((((.((((.......))))..))))))((((...)))).((((....((((((.........))))))......))))... ( -22.70) >DroSec_CAF1 18830 90 + 1 AAACCCGCACAAAGCCGGAAAACGUGGCAAUUGUGUGCUGUGCCAGCAUCUCAAACCCAUGUCCUCCUUCCACUUUGCACCUGGAGAAAA ....(((..(((((..(((..((((((...(((.((((((...))))))..)))..))))))..))).....)))))....)))...... ( -21.90) >DroSim_CAF1 20072 90 + 1 AAACCCGCACAAAGCCGGAAAACGUGGCAAUUGUGUGCUGGCCCAGCAUCUCAAACCCAUGUCCUCCUACUACUUUGCACUUGGAGAAAA ....(((..(((((..(((..((((((...(((.((((((...))))))..)))..))))))..))).....)))))....)))...... ( -21.90) >DroEre_CAF1 14155 90 + 1 AAACCCGCACAAAGCCGGAAAACGUGGCAAUUGAGUGCCGGCCCAGCAUCUCAAACCCACGUCCUCCUCUUACUGUGCACCUGGAGAAAA ......((((((((..(((..((((((...((((((((.......))).)))))..))))))..))).)))..)))))............ ( -28.00) >DroYak_CAF1 17110 90 + 1 AAACCCGCACAAAGCCGGAAAACGUGGCAAUUGUGUGCCGGCCCAGCAUCUCAAAACCAUGUCCUCCCUCUACUGUGCACCUGGAGAAAA ......(((((.((..(((..((((((...(((.((((.......))))..)))..))))))..)))..))..)))))............ ( -22.60) >consensus AAACCCGCACAAAGCCGGAAAACGUGGCAAUUGUGUGCUGGCCCAGCAUCUCAAACCCAUGUCCUCCUUCUACUUUGCACCUGGAGAAAA ...((.((.....)).))...((((((...(((.((((((...))))))..)))..)))))).((((...............)))).... (-19.32 = -19.56 + 0.24)

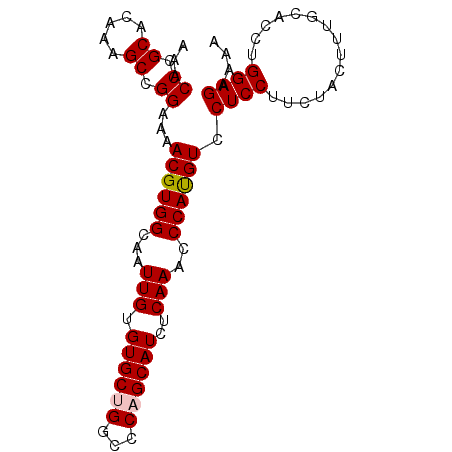
| Location | 4,264,194 – 4,264,284 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -31.26 |
| Consensus MFE | -28.72 |
| Energy contribution | -29.28 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4264194 90 - 22407834 UUUUCUCCAGGUGCCAUGUAGAAGGAGGACAUGGGUUUGAGAUGCUGGGCCAGCACACAAUUGCCACGUUUUCCGGCUUUGUGCGGGUUU ......((..(..(...((....(((((((.(((..(((.(.(((((...)))))).)))...))).))))))).))...)..).))... ( -24.80) >DroSec_CAF1 18830 90 - 1 UUUUCUCCAGGUGCAAAGUGGAAGGAGGACAUGGGUUUGAGAUGCUGGCACAGCACACAAUUGCCACGUUUUCCGGCUUUGUGCGGGUUU ......((..(..((((((....(((((((.(((..(((.(.(((((...)))))).)))...))).))))))).))))))..).))... ( -31.00) >DroSim_CAF1 20072 90 - 1 UUUUCUCCAAGUGCAAAGUAGUAGGAGGACAUGGGUUUGAGAUGCUGGGCCAGCACACAAUUGCCACGUUUUCCGGCUUUGUGCGGGUUU ......((..(..((((((....(((((((.(((..(((.(.(((((...)))))).)))...))).))))))).))))))..).))... ( -31.20) >DroEre_CAF1 14155 90 - 1 UUUUCUCCAGGUGCACAGUAAGAGGAGGACGUGGGUUUGAGAUGCUGGGCCGGCACUCAAUUGCCACGUUUUCCGGCUUUGUGCGGGUUU ......((..(..((.(((....(((((((((((..(((((.(((((...))))))))))...))))))))))).))).))..).))... ( -38.50) >DroYak_CAF1 17110 90 - 1 UUUUCUCCAGGUGCACAGUAGAGGGAGGACAUGGUUUUGAGAUGCUGGGCCGGCACACAAUUGCCACGUUUUCCGGCUUUGUGCGGGUUU ...........(((((((.((..(((((((.((((.(((.(.(((((...)))))).)))..)))).)))))))..)))))))))..... ( -30.80) >consensus UUUUCUCCAGGUGCAAAGUAGAAGGAGGACAUGGGUUUGAGAUGCUGGGCCAGCACACAAUUGCCACGUUUUCCGGCUUUGUGCGGGUUU ......((..(..((((((....(((((((.(((..(((.(.(((((...)))))).)))...))).))))))).))))))..).))... (-28.72 = -29.28 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:02 2006