| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,257,149 – 4,257,246 |
| Length | 97 |
| Max. P | 0.784446 |

| Location | 4,257,149 – 4,257,246 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -16.09 |
| Energy contribution | -16.48 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
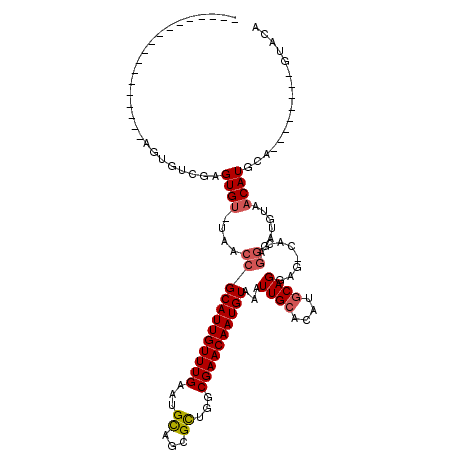
>2L_DroMel_CAF1 4257149 97 - 22407834 -----------GUGUG--UGUGUGGAGUGU-UAACCGCAUUGUUUGAAUGCAGCGCUGGCGAACAAUGUACAUUGCACAUGCAGGAG-CACAGGGAUGUAACAUGCA--------GUACA -----------(((((--((((((.(((((-.....((((((((((...((...))...))))))))))))))).)))))))....)-))).(...(((.....)))--------...). ( -29.60) >DroSec_CAF1 11895 91 - 1 ------------------AGUGUGGAGUGU-UAACCGCAUUGUUUGAAUGUAGCGCU-GCGAACAAUGUACAUUGCACAUGCAGGAG-CACAGGGAUGUAACAUGCA--------GUACA ------------------.(((((.(((((-.....((((((((((...(.....).-.))))))))))))))).)))))((((..(-((......)))..).))).--------..... ( -25.90) >DroSim_CAF1 13084 92 - 1 ------------------AGUGUGGAGUGU-UAACCGCAUUGUUUGAAUGCAGCGCUGGCGAACAAUGUACAUUGCACAUGCAGGAG-CACAGGGAUGUAACAUUCA--------GUACA ------------------.(((..((((((-((.((((((((((((...((...))...))))))))))...((((....))))...-....))....)))))))).--------.))). ( -29.80) >DroEre_CAF1 7159 92 - 1 ------------------AGUGUCGAGUGU-UAACCGCAUUGUUUGAAUGCACUGCUGGCGAACAAUGUGAAUUGCACAUGCAGGAG-CCCAGGGAUGUAACAUGCA--------GUACA ------------------.(((..(.((((-((.(((((((.....))))).(((..(((.....(((((.....)))))......)-)))))))...)))))).).--------.))). ( -26.10) >DroYak_CAF1 9893 101 - 1 ------------------AGUGUCGAGUGU-UAACCGCAUUGUUUGAAUGCACUGCUGGCGAACAAUGUGAAUUGCACAUGCAGGAGGCACAGGGAUGUAACAUGUACACUACAUGUACA ------------------.(((((......-....(((((((((((...((...))...)))))))))))..((((....))))..))))).....((((.((((((...)))))))))) ( -31.60) >DroPer_CAF1 27737 100 - 1 AGAGUGUACGUCUGUGUGAGUG-CGAGUGCACAACCGCAUUGUUUGAAUGCUGCGUUGGCGAACAAUGUAAAUUGC-----CAGCAG-UACU---AUAU-ACAUA-U--------GUACA ....(((((((.((((((((((-(....(((.....(((((.....))))))))((((((((..........))))-----)))).)-))))---.)))-))).)-)--------))))) ( -34.60) >consensus __________________AGUGUCGAGUGU_UAACCGCAUUGUUUGAAUGCAGCGCUGGCGAACAAUGUAAAUUGCACAUGCAGGAG_CACAGGGAUGUAACAUGCA________GUACA ..........................((((....((((((((((((...((...))...))))))))))...((((....))))........))......))))................ (-16.09 = -16.48 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:57 2006