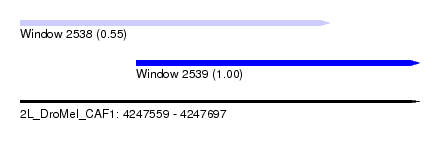
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,247,559 – 4,247,697 |
| Length | 138 |
| Max. P | 0.999646 |

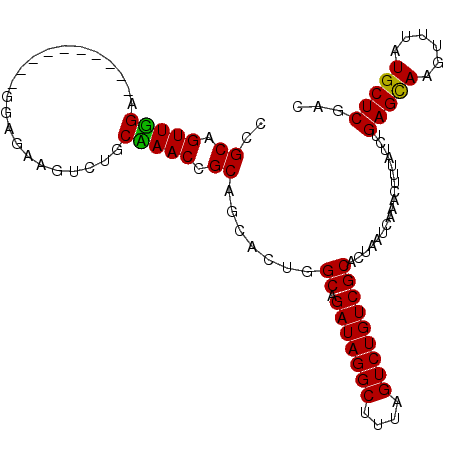
| Location | 4,247,559 – 4,247,666 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 86.50 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -23.12 |
| Energy contribution | -22.35 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4247559 107 + 22407834 CCGCAGUUGGAGCUCAAGUUUGGGGAUGUCUGCUAACCGCCGCGCUGGCAGAUAGGCUUUAGUCUGUCGCACUAAUCAAACUUUAUCUGAGCAAGUUUAUGCUCUAC ..(((......(((((..(..((.(((...(((.....(((.....))).(((((((....))))))))))...)))...))..)..))))).......)))..... ( -31.12) >DroEre_CAF1 25756 97 + 1 CCGCAGUUGGU----------GGAGAAGUCUGCCAACCGCGGCACUGGCAGAUAGGCUUUAGUCUGUCGCACUAAUCAAACUUUAUCUGAGCAAGUUUAUGCUCGAC ((((.(((((.----------.((....))..))))).)))).....((.(((((((....))))))))).................((((((......)))))).. ( -36.40) >DroYak_CAF1 31536 97 + 1 CCGCAGUUUGA----------GGAGAAGUCUGCAAACCGCAGCACUGGCUGAUAGGCUUUAGUCUGUCGCACUAAUCAAACUUUAUCUGAGUAAGUUUAUGCUCGAC ....(((((((----------..((.(((((((.....)))).))).((.(((((((....))))))))).))..))))))).....((((((......)))))).. ( -31.40) >consensus CCGCAGUUGGA__________GGAGAAGUCUGCAAACCGCAGCACUGGCAGAUAGGCUUUAGUCUGUCGCACUAAUCAAACUUUAUCUGAGCAAGUUUAUGCUCGAC ..((.(((((......................))))).)).......((.(((((((....)))))))))..................(((((......)))))... (-23.12 = -22.35 + -0.77)

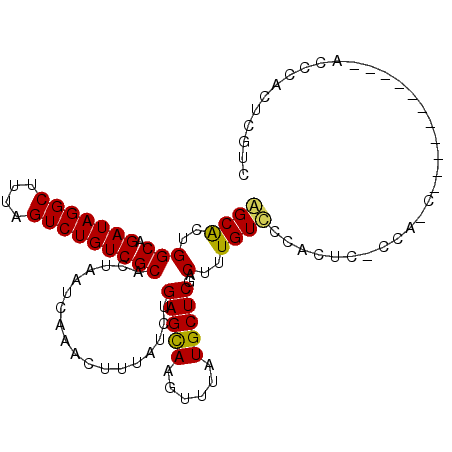
| Location | 4,247,599 – 4,247,697 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 73.40 |
| Mean single sequence MFE | -18.47 |
| Consensus MFE | -18.13 |
| Energy contribution | -17.80 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4247599 98 + 22407834 CGCGCUGGCAGAUAGGCUUUAGUCUGUCGCACUAAUCAAACUUUAUCUGAGCAAGUUUAUGCUCUACUUUGUCCCACUCCCCAACA---------UACCCACUCGUC .(((.(((..(((((((....)))))))....................(((((......)))))......................---------...)))..))). ( -18.30) >DroEre_CAF1 25786 107 + 1 GGCACUGGCAGAUAGGCUUUAGUCUGUCGCACUAAUCAAACUUUAUCUGAGCAAGUUUAUGCUCGACUUUGUCCCACUCACCACCUACUACUCACCACCCACUCGUC ((((..(((.(((((((....))))))))).................((((((......)))))).)..)))).................................. ( -20.60) >DroYak_CAF1 31566 74 + 1 AGCACUGGCUGAUAGGCUUUAGUCUGUCGCACUAAUCAAACUUUAUCUGAGUAAGUUUAUGCUCGACUUUGUUC--------------------------------- ((((..(((((((((((....)))))))...........((((.....)))).))))..))))...........--------------------------------- ( -16.50) >consensus AGCACUGGCAGAUAGGCUUUAGUCUGUCGCACUAAUCAAACUUUAUCUGAGCAAGUUUAUGCUCGACUUUGUCCCACUC_CCA_C___________ACCCACUCGUC ((((..(((.(((((((....)))))))))..................(((((......)))))..)..)))).................................. (-18.13 = -17.80 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:51 2006