| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,243,945 – 4,244,050 |
| Length | 105 |
| Max. P | 0.892649 |

| Location | 4,243,945 – 4,244,050 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 88.73 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -21.42 |
| Energy contribution | -22.18 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4243945 105 + 22407834 ACGAUGUGGCACUGCACGUUGCAUACAUGGUAGUAACCAUUAAUGCAACAUCAUGGGGGGAAAUACAUUUGCAAGGAAACUUCUUCUUCGAUGCAUAAAAACGUA ...((((.....((((((((((((..(((((....)))))..))))))).......((((((........(.(((....))))))))))).)))).....)))). ( -25.50) >DroSec_CAF1 18974 105 + 1 GCGAUGUGGCACUGCACGUUGCAUACAUGGUAGUAACCAUUAAUGCAACAUCACGGGGGGAAAUACAUUUGCAAGGAAACUUCUUCUUCGAUGCAUUAAAAUUUA ((((((((......)))(((((((..(((((....)))))..)))))))))).((((((.(((....))).)(((....)))...)))))..))........... ( -26.40) >DroEre_CAF1 22148 105 + 1 GCGAUGUGGCACAGAGCGUUGCAUACAUGGUUGUAACCAUUAAUGCAACAUCACGGGGGGAAAUACAUUUGCAAGGAAACUUCUUCGUCGAUGCAUAAAAACGUA ((..((.....))..))(((((((..(((((....)))))..)))))))((((((((((.(((....))).)(((....))))))))).)))............. ( -27.20) >DroYak_CAF1 27774 96 + 1 GA---------GAGCGUGUUGCAUACAUGGUAGUAACCAUUAAUGCAACAUCACGGGGGGAAAUACAUUUGCAAGGAAACUUCUUCGUCGAUGCAUGAAAACGUA ..---------..(((((((((((..(((((....)))))..)))))))((((((((((.(((....))).)(((....))))))))).)))........)))). ( -26.90) >consensus GCGAUGUGGCACAGCACGUUGCAUACAUGGUAGUAACCAUUAAUGCAACAUCACGGGGGGAAAUACAUUUGCAAGGAAACUUCUUCGUCGAUGCAUAAAAACGUA .............((..(((((((..(((((....)))))..)))))))((((((((((.(((....))).)(((....))))))))).)))))........... (-21.42 = -22.18 + 0.75)


| Location | 4,243,945 – 4,244,050 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 88.73 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -20.70 |
| Energy contribution | -21.08 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
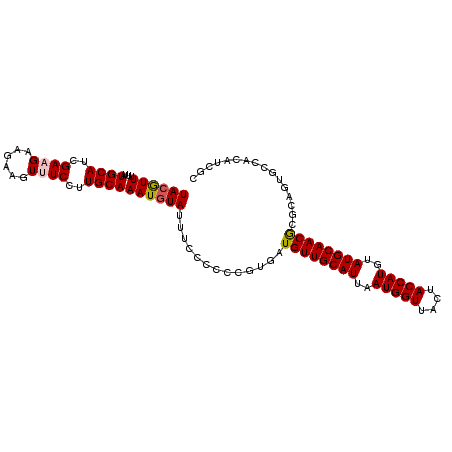
>2L_DroMel_CAF1 4243945 105 - 22407834 UACGUUUUUAUGCAUCGAAGAAGAAGUUUCCUUGCAAAUGUAUUUCCCCCCAUGAUGUUGCAUUAAUGGUUACUACCAUGUAUGCAACGUGCAGUGCCACAUCGU ((((((....((((..((((......))))..)))))))))).........(((((((.(((((.(((((....)))))(((((...)))))))))).))))))) ( -27.40) >DroSec_CAF1 18974 105 - 1 UAAAUUUUAAUGCAUCGAAGAAGAAGUUUCCUUGCAAAUGUAUUUCCCCCCGUGAUGUUGCAUUAAUGGUUACUACCAUGUAUGCAACGUGCAGUGCCACAUCGC ..........((((..((((......))))..))))...............(((((((.(((((.(((((....)))))(((((...)))))))))).))))))) ( -26.90) >DroEre_CAF1 22148 105 - 1 UACGUUUUUAUGCAUCGACGAAGAAGUUUCCUUGCAAAUGUAUUUCCCCCCGUGAUGUUGCAUUAAUGGUUACAACCAUGUAUGCAACGCUCUGUGCCACAUCGC ((((((....((((..(((......)))....)))))))))).........((((((((((((..(((((....)))))..)))))))((.....))...))))) ( -25.60) >DroYak_CAF1 27774 96 - 1 UACGUUUUCAUGCAUCGACGAAGAAGUUUCCUUGCAAAUGUAUUUCCCCCCGUGAUGUUGCAUUAAUGGUUACUACCAUGUAUGCAACACGCUC---------UC ((((((....((((..(((......)))....)))))))))).........(((.((((((((..(((((....)))))..)))))))))))..---------.. ( -25.80) >consensus UACGUUUUUAUGCAUCGAAGAAGAAGUUUCCUUGCAAAUGUAUUUCCCCCCGUGAUGUUGCAUUAAUGGUUACUACCAUGUAUGCAACGCGCAGUGCCACAUCGC ((((((....((((..((((......))))..)))))))))).............((((((((..(((((....)))))..))))))))................ (-20.70 = -21.08 + 0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:50 2006