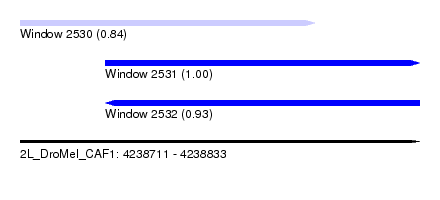
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,238,711 – 4,238,833 |
| Length | 122 |
| Max. P | 0.996673 |

| Location | 4,238,711 – 4,238,801 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 92.11 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -20.94 |
| Energy contribution | -21.00 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
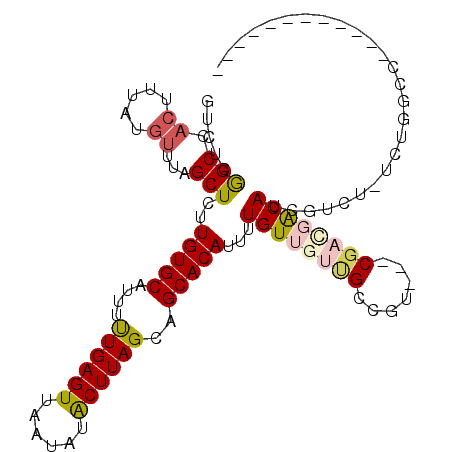
>2L_DroMel_CAF1 4238711 90 + 22407834 U------CUGCUCUUCCUUUCGUCCUUUCUCGGGCCAGUAGACGAUGUCGUCGGGGACAACAACAAAAUGUGCUGCUAAGCAUAUUAACUCAAAAA .------.((.(((((.....((((......)))).....((((....))))))))))).......((((((((....)))))))).......... ( -22.00) >DroSec_CAF1 13808 90 + 1 U------CUGCUCUUCCUUUCGUCCUUUCUCGGGCCAGCAGACGAUGUCGUCGGGGACAACAACAAAAUGUGCUGCUAAGCAUAUUAACUCAAAAA (------(((((.........((((......)))).))))))...(((.((.(....).)).))).((((((((....)))))))).......... ( -22.20) >DroSim_CAF1 20955 90 + 1 U------CUGCUCUUUCUUUCGUCCUUUCUCGGGCCAGCAGACGAUGUCGUCGGGGACAACAACAAAAUGUGCUGCUAAGCAUAUUAACUCAAAAA (------(((((.........((((......)))).))))))...(((.((.(....).)).))).((((((((....)))))))).......... ( -22.20) >DroYak_CAF1 22267 93 + 1 UGCUGUCCUGCUCUUCUUUUCGUCCUUUCUUGGGCCAGAAGACGAUGUCGUCGUGGACA---ACAAAAUGUGCUGCUAAGCAUAUUAACUCAAAAA ((.(((((.((.....((((.((((......)))).))))((((....)))))))))))---.)).((((((((....)))))))).......... ( -28.00) >consensus U______CUGCUCUUCCUUUCGUCCUUUCUCGGGCCAGCAGACGAUGUCGUCGGGGACAACAACAAAAUGUGCUGCUAAGCAUAUUAACUCAAAAA ........((.(((((.....((((......)))).....((((....))))))))))).......((((((((....)))))))).......... (-20.94 = -21.00 + 0.06)


| Location | 4,238,737 – 4,238,833 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 74.06 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -10.87 |
| Energy contribution | -11.03 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.39 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4238737 96 + 22407834 ------------GGCCAGU-AGACGAUGUCGUCG---GGGACAACAACAAAAUGUGCUGCUAAGCAUAUUAACUCAAAAAUGCACAAAAGCAAAACAUAAAGUGGCCAAGAC ------------(((((.(-.((((....))))(---....)........((((((((....))))))))..........(((......)))........).)))))..... ( -27.00) >DroVir_CAF1 12728 111 + 1 GCGGCAAGCGCCGGACAGC-UGAGGAUGUCGUCGUCAACGGCGACAACAAAAUGUGCUGCUAAGUAUAUUAACUCAGAAAUGCACAAGAGCUAAACAUAAAGUAGCCGGCCA ((.....))(((((.(..(-((((..((((((((....))))))))....((((((((....))))))))..)))))....((......)).............)))))).. ( -38.70) >DroGri_CAF1 18255 106 + 1 --AGAAGGAGAAGGA-GGA-CGAAGAUGCCAUCGUCAACGGCUACAACAAAAUGUGCUGCUAAGUAUAUUAACUCAAAAAUGCACAAGAGCUAAACAUCU--GAGCUGAUUG --.............-.((-(((.(....).)))))..(((((.((....((((((((....))))))))...........((......))........)--)))))).... ( -20.00) >DroEre_CAF1 14249 94 + 1 ------------GGCCAGAAAGACGAUGUCGUCG---GCGACA---ACAAAAUGUGCUGCUAAGCAUAUUAACUCAAAAAUGCACAAAAGCAAAACAUAAAGUGGCCAAGAC ------------(((((((.....(.(((((...---.)))))---.)..((((((((....))))))))...)).....(((......)))..........)))))..... ( -26.90) >DroYak_CAF1 22299 93 + 1 ------------GGCCAGA-AGACGAUGUCGUCG---UGGACA---ACAAAAUGUGCUGCUAAGCAUAUUAACUCAAAAAUGCACAAAAGCAAAACAUAAAGUGGCCAAGAC ------------(((((..-.((((....))))(---((....---....((((((((....))))))))..........(((......)))...)))....)))))..... ( -26.80) >DroMoj_CAF1 12562 93 + 1 ------------------C-UGUGGAUGUCGUCGUCAACGGCGACAACAAAAUGUGCUGCUAAGUAUAUUAACUCAGAAAUGCACAAGAGCUAAACAUAAAGUAGCUGGCCG ------------------.-...((.((((((((....))))))))......(((((..((.(((......))).))....)))))..(((((.........)))))..)). ( -26.10) >consensus ____________GGCCAGA_AGACGAUGUCGUCG___ACGACAACAACAAAAUGUGCUGCUAAGCAUAUUAACUCAAAAAUGCACAAAAGCAAAACAUAAAGUAGCCAAGAC .........................((((.((((....))))........((((((((....))))))))...........((......))...)))).............. (-10.87 = -11.03 + 0.17)

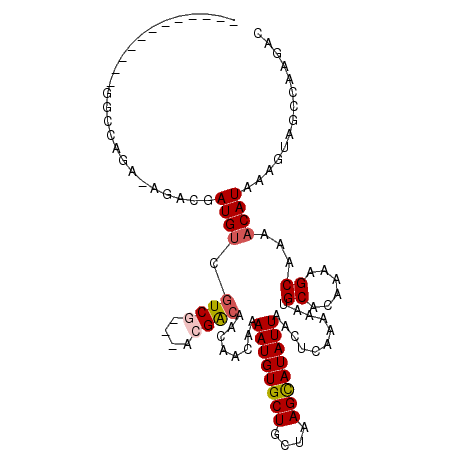

| Location | 4,238,737 – 4,238,833 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 74.06 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.93 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4238737 96 - 22407834 GUCUUGGCCACUUUAUGUUUUGCUUUUGUGCAUUUUUGAGUUAAUAUGCUUAGCAGCACAUUUUGUUGUUGUCCC---CGACGACAUCGUCU-ACUGGCC------------ .....(((((....((((.(((((.....((((.((......)).))))..))))).))))..((((((((....---))))))))......-..)))))------------ ( -25.30) >DroVir_CAF1 12728 111 - 1 UGGCCGGCUACUUUAUGUUUAGCUCUUGUGCAUUUCUGAGUUAAUAUACUUAGCAGCACAUUUUGUUGUCGCCGUUGACGACGACAUCCUCA-GCUGUCCGGCGCUUGCCGC .(((.(((.......((..(((((..(((((....((((((......))))))..)))))...((((((((.......)))))))).....)-))))..))..))).))).. ( -34.50) >DroGri_CAF1 18255 106 - 1 CAAUCAGCUC--AGAUGUUUAGCUCUUGUGCAUUUUUGAGUUAAUAUACUUAGCAGCACAUUUUGUUGUAGCCGUUGACGAUGGCAUCUUCG-UCC-UCCUUCUCCUUCU-- ..........--.((((..((((...(((((....((((((......))))))..)))))....))))..((((((...)))))).....))-)).-.............-- ( -19.90) >DroEre_CAF1 14249 94 - 1 GUCUUGGCCACUUUAUGUUUUGCUUUUGUGCAUUUUUGAGUUAAUAUGCUUAGCAGCACAUUUUGU---UGUCGC---CGACGACAUCGUCUUUCUGGCC------------ .....(((((....((((.(((((.....((((.((......)).))))..))))).))))..(((---((((..---.))))))).........)))))------------ ( -23.90) >DroYak_CAF1 22299 93 - 1 GUCUUGGCCACUUUAUGUUUUGCUUUUGUGCAUUUUUGAGUUAAUAUGCUUAGCAGCACAUUUUGU---UGUCCA---CGACGACAUCGUCU-UCUGGCC------------ .....(((((....((((.(((((.....((((.((......)).))))..))))).))))..(((---((((..---.)))))))......-..)))))------------ ( -23.90) >DroMoj_CAF1 12562 93 - 1 CGGCCAGCUACUUUAUGUUUAGCUCUUGUGCAUUUCUGAGUUAAUAUACUUAGCAGCACAUUUUGUUGUCGCCGUUGACGACGACAUCCACA-G------------------ .((..(((((.........)))))..(((((....((((((......))))))..)))))...((((((((.......)))))))).))...-.------------------ ( -24.10) >consensus GUCUCGGCCACUUUAUGUUUAGCUCUUGUGCAUUUUUGAGUUAAUAUACUUAGCAGCACAUUUUGUUGUUGCCGU___CGACGACAUCGUCU_UCUGGCC____________ .....(((.((.....))...)))..(((((....((((((......))))))..)))))...((((((((.......)))))))).......................... (-14.52 = -14.93 + 0.42)
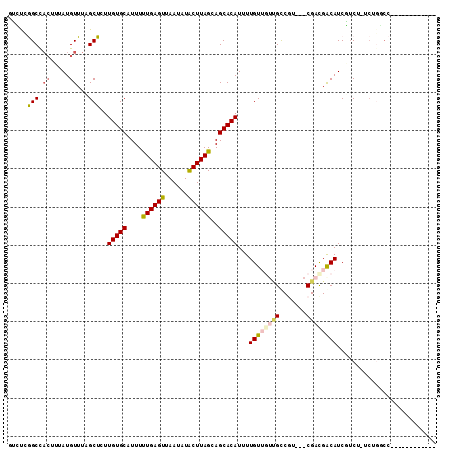
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:45 2006